Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
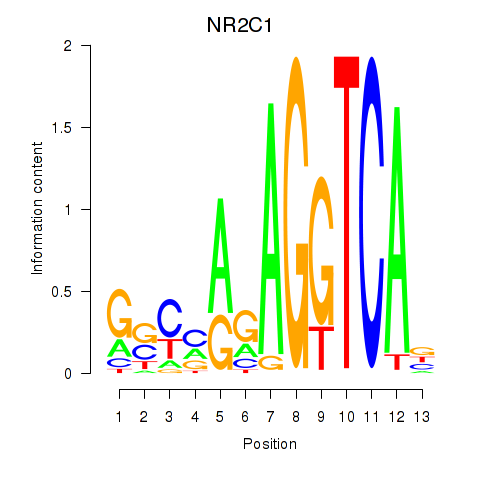
Results for NR2C1
Z-value: 1.59
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.12 | nuclear receptor subfamily 2 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C1 | hg19_v2_chr12_-_95467267_95467350 | 0.04 | 8.2e-01 | Click! |
Activity profile of NR2C1 motif
Sorted Z-values of NR2C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_229569834 | 8.51 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr17_+_37821593 | 6.33 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr17_+_4853442 | 5.65 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr16_-_4292071 | 5.52 |
ENST00000399609.3
|
SRL
|
sarcalumenin |
| chr1_-_917466 | 5.36 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr9_-_35685452 | 4.95 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr14_-_65289812 | 4.61 |
ENST00000389720.3
ENST00000389721.5 ENST00000389722.3 |
SPTB
|
spectrin, beta, erythrocytic |
| chr1_+_160097462 | 4.48 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr14_+_105212297 | 4.45 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr12_+_48499883 | 4.38 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr22_+_31518938 | 4.24 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr18_+_7231123 | 4.05 |
ENST00000383467.2
|
LRRC30
|
leucine rich repeat containing 30 |
| chrX_+_38420623 | 3.82 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr6_+_133561725 | 3.77 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chrX_+_38420783 | 3.54 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr4_-_111544254 | 3.48 |
ENST00000306732.3
|
PITX2
|
paired-like homeodomain 2 |
| chr22_-_50699701 | 3.40 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr19_-_48867171 | 3.32 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr19_-_48867291 | 3.17 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr19_+_16771936 | 3.04 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr4_-_111544207 | 2.90 |
ENST00000557119.2
|
PITX2
|
paired-like homeodomain 2 |
| chr15_-_43863695 | 2.85 |
ENST00000439195.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr19_-_51289436 | 2.75 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr6_-_160148356 | 2.50 |
ENST00000401980.3
ENST00000545162.1 |
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr3_+_37284824 | 2.49 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr3_-_58613323 | 2.38 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr3_+_159557637 | 2.36 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_+_24755416 | 2.33 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr14_-_73360796 | 2.14 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr11_+_94501497 | 2.12 |
ENST00000317829.8
ENST00000317837.9 ENST00000433060.2 |
AMOTL1
|
angiomotin like 1 |
| chr15_+_65369082 | 2.11 |
ENST00000432196.2
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr6_-_160147925 | 1.95 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr10_-_118502070 | 1.88 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr8_-_80680078 | 1.87 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr15_-_83474806 | 1.79 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr15_-_65715401 | 1.78 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr12_+_109577202 | 1.78 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr17_+_13972807 | 1.72 |
ENST00000429152.2
ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10
|
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr9_-_33402506 | 1.72 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr10_+_63808970 | 1.72 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr13_-_33760216 | 1.67 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_+_64073699 | 1.64 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr11_+_131781290 | 1.62 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr8_+_96037205 | 1.59 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_-_163099546 | 1.49 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr1_+_44412577 | 1.49 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr3_+_37284668 | 1.49 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr7_-_229557 | 1.45 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr11_+_10471836 | 1.39 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr14_-_21492251 | 1.36 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr6_+_31515337 | 1.30 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr2_+_202316392 | 1.27 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr8_-_145028013 | 1.27 |
ENST00000354958.2
|
PLEC
|
plectin |
| chr22_-_38851205 | 1.26 |
ENST00000303592.3
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr5_+_1801503 | 1.24 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr8_+_96037255 | 1.24 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_-_163099885 | 1.22 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr4_+_159593418 | 1.21 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr11_+_111896090 | 1.11 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr3_-_116164306 | 1.06 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_+_215255136 | 1.06 |
ENST00000478774.1
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr15_+_91449971 | 1.06 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr19_+_51293672 | 1.06 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr13_+_103046954 | 1.04 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr2_-_163100045 | 1.03 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr11_+_111896320 | 0.98 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr14_-_21492113 | 0.96 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_49371711 | 0.94 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr11_+_67798090 | 0.91 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr3_-_50605150 | 0.90 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr12_-_124873357 | 0.89 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr8_+_70404996 | 0.87 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chrX_-_128657457 | 0.87 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr15_+_82555125 | 0.81 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr18_+_8609402 | 0.78 |
ENST00000329286.6
|
RAB12
|
RAB12, member RAS oncogene family |
| chr10_-_23633720 | 0.77 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr3_+_46538981 | 0.71 |
ENST00000296142.3
|
RTP3
|
receptor (chemosensory) transporter protein 3 |
| chr11_-_64013288 | 0.70 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr20_-_30458432 | 0.70 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr20_-_44485835 | 0.69 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr22_+_39745930 | 0.69 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr2_+_109237717 | 0.69 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr10_+_104005272 | 0.66 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr6_+_43737939 | 0.64 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr19_-_51472031 | 0.62 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr22_+_41777927 | 0.61 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr8_-_80942467 | 0.57 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr3_+_154801312 | 0.57 |
ENST00000497890.1
|
MME
|
membrane metallo-endopeptidase |
| chr3_-_50605077 | 0.57 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr3_+_150264555 | 0.56 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chrX_-_132095419 | 0.53 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_+_45140400 | 0.53 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr3_+_154801678 | 0.51 |
ENST00000462837.1
|
MME
|
membrane metallo-endopeptidase |
| chr2_-_219134343 | 0.51 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr3_+_184055240 | 0.51 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr9_-_139839064 | 0.51 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr3_+_150264458 | 0.50 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr11_-_64013663 | 0.50 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_205419053 | 0.49 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr6_+_160148593 | 0.46 |
ENST00000337387.4
|
WTAP
|
Wilms tumor 1 associated protein |
| chr2_+_151485403 | 0.45 |
ENST00000413247.1
ENST00000423428.1 ENST00000427615.1 ENST00000443811.1 |
AC104777.4
|
AC104777.4 |
| chr2_+_201936458 | 0.45 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr18_-_54318353 | 0.45 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr2_+_210636697 | 0.45 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr11_+_7595136 | 0.44 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr13_+_21714653 | 0.42 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr16_-_88752889 | 0.42 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr2_+_201936707 | 0.42 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr16_+_83932684 | 0.42 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr22_-_31536480 | 0.41 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr19_-_41256207 | 0.40 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr22_+_30163340 | 0.39 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr9_-_16253112 | 0.37 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr11_-_26743546 | 0.36 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_-_226374373 | 0.35 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr3_-_116163830 | 0.35 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_-_65429891 | 0.35 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr20_+_19870167 | 0.35 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_+_57037089 | 0.34 |
ENST00000370693.5
|
BAG2
|
BCL2-associated athanogene 2 |
| chr15_-_82555000 | 0.34 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr14_+_74035763 | 0.32 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr2_+_109745655 | 0.32 |
ENST00000418513.1
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr1_-_120439112 | 0.31 |
ENST00000369400.1
|
ADAM30
|
ADAM metallopeptidase domain 30 |
| chr13_+_21714711 | 0.28 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr5_-_42811986 | 0.27 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_+_67798363 | 0.27 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr15_+_45422178 | 0.26 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr15_+_45422131 | 0.26 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr6_+_10585979 | 0.24 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr5_-_1801408 | 0.24 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr11_+_67798114 | 0.24 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr22_+_47158578 | 0.23 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr19_-_36054555 | 0.22 |
ENST00000262623.3
|
ATP4A
|
ATPase, H+/K+ exchanging, alpha polypeptide |
| chr4_-_76598296 | 0.21 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr15_-_45422056 | 0.20 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr11_+_123810250 | 0.18 |
ENST00000307033.2
|
OR4D5
|
olfactory receptor, family 4, subfamily D, member 5 |
| chr2_-_96701722 | 0.18 |
ENST00000434632.1
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr7_+_142749417 | 0.16 |
ENST00000418316.1
|
OR6V1
|
olfactory receptor, family 6, subfamily V, member 1 |
| chr16_+_2255710 | 0.15 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_-_45140074 | 0.15 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr9_-_34397800 | 0.15 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr12_-_109219937 | 0.15 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr4_-_76598326 | 0.14 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr9_+_33795533 | 0.13 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr14_+_74003818 | 0.12 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr12_-_54070098 | 0.12 |
ENST00000394349.3
ENST00000549164.1 |
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr10_+_127661942 | 0.10 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr9_-_140513231 | 0.09 |
ENST00000371417.3
|
C9orf37
|
chromosome 9 open reading frame 37 |
| chr1_+_212797789 | 0.09 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr16_+_2255841 | 0.08 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr7_+_121513143 | 0.08 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr19_-_46142362 | 0.07 |
ENST00000586770.1
ENST00000591721.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr12_-_53729525 | 0.06 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr3_+_179065474 | 0.06 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr12_-_42631529 | 0.05 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr13_+_21714913 | 0.05 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr16_-_70729496 | 0.04 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr11_-_63684316 | 0.04 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr3_+_183903811 | 0.02 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr2_-_219134822 | 0.01 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr2_+_46926326 | 0.01 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_+_33701707 | 0.01 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_219433014 | 0.00 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 1.4 | 8.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.3 | 6.4 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 1.2 | 3.7 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.9 | 6.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.9 | 4.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.9 | 4.4 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.7 | 4.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.7 | 2.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 1.9 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.3 | 2.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 1.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.6 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 1.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 3.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 1.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 0.7 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 3.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 2.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 4.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 4.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 2.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 2.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 5.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 6.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.7 | GO:0008535 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 5.0 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 4.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 1.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.7 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 1.3 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 1.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.7 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 2.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 4.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 2.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 5.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 5.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 8.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 4.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 3.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 3.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 4.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 5.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 1.1 | 6.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.7 | 2.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.6 | 5.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 4.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 1.9 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 4.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 1.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 2.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 1.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 4.2 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 1.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 4.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 1.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.2 | 1.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 5.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 8.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 3.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 3.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 3.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 6.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 3.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 5.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 1.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 6.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 8.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 4.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 10.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 11.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 4.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 4.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |