Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
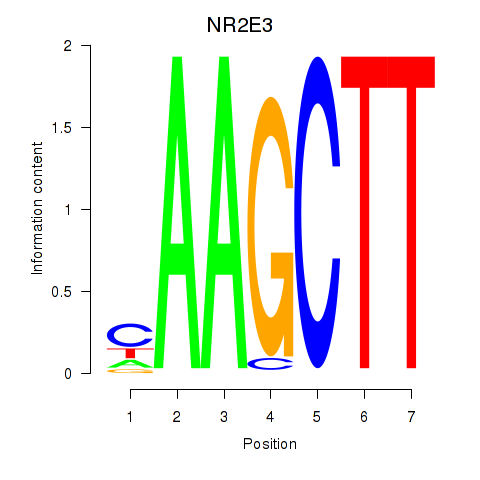
Results for NR2E3
Z-value: 2.17
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_138210977 | 7.02 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr5_-_138211051 | 5.85 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr15_+_54305101 | 4.97 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr9_+_72002837 | 4.51 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr3_-_195310802 | 4.43 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr1_-_102312517 | 4.43 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr9_-_93405352 | 4.10 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr12_-_99548645 | 3.68 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_164924716 | 3.59 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr2_+_166095898 | 3.52 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr3_-_58563094 | 3.45 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr3_+_63428982 | 3.45 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr6_+_53883708 | 3.38 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr1_+_66999799 | 3.38 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr1_-_102312600 | 3.31 |
ENST00000359814.3
|
OLFM3
|
olfactomedin 3 |
| chr9_+_74764340 | 3.18 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr1_-_144995074 | 3.13 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr16_+_7382745 | 3.11 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr22_+_41865109 | 3.10 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr4_+_88896819 | 3.06 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr3_+_181429704 | 3.05 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr5_+_173473662 | 2.90 |
ENST00000519717.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr3_+_63428752 | 2.85 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr12_+_79258444 | 2.77 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr14_+_32964258 | 2.69 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_-_124084493 | 2.64 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr14_+_78870030 | 2.60 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr12_+_41221794 | 2.60 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr16_-_75529273 | 2.58 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr18_-_40857447 | 2.44 |
ENST00000590752.1
ENST00000596867.1 |
SYT4
|
synaptotagmin IV |
| chr18_-_72125179 | 2.38 |
ENST00000400291.2
|
FAM69C
|
family with sequence similarity 69, member C |
| chr4_-_87374283 | 2.35 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_161275320 | 2.32 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_+_155555201 | 2.32 |
ENST00000544049.1
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr9_+_74764278 | 2.32 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr1_+_154540246 | 2.26 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr1_-_144995002 | 2.22 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr5_-_87980753 | 2.21 |
ENST00000511014.2
|
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr2_-_175712270 | 2.17 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr9_+_33795533 | 2.17 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr2_-_175712479 | 2.16 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr10_-_103603568 | 2.16 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr9_+_71986182 | 2.16 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_20848377 | 2.15 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr10_-_21186144 | 2.12 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr6_+_69942298 | 2.12 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr2_-_50201327 | 2.12 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr11_-_45307817 | 2.12 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr5_+_137774706 | 2.10 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chrX_-_13835398 | 2.07 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr15_+_42696954 | 2.07 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr10_-_93392811 | 2.07 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr7_-_137028498 | 2.06 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chrX_-_13835147 | 2.06 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr2_-_224467093 | 2.05 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr9_+_71939488 | 2.04 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr11_-_88796803 | 2.04 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr7_-_137028534 | 2.03 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr15_+_42697065 | 2.03 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr5_-_87980587 | 2.02 |
ENST00000509783.1
ENST00000509405.1 ENST00000506978.1 ENST00000509265.1 ENST00000513805.1 |
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr4_-_20985632 | 2.02 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr1_+_65775204 | 2.00 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr6_+_53883790 | 2.00 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr7_-_79082867 | 1.99 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_161274685 | 1.97 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr12_+_20848282 | 1.97 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr3_+_164924769 | 1.96 |
ENST00000494915.1
|
RP11-85M11.2
|
RP11-85M11.2 |
| chr5_-_11589131 | 1.94 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr9_+_71944241 | 1.94 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_161274940 | 1.94 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_-_186732892 | 1.91 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_33686925 | 1.91 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr15_+_42696992 | 1.91 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr2_-_175711978 | 1.90 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr11_-_40315640 | 1.90 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr7_-_79082786 | 1.88 |
ENST00000522391.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_114214125 | 1.87 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr12_+_79258547 | 1.85 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr18_-_40857329 | 1.85 |
ENST00000593720.1
|
SYT4
|
synaptotagmin IV |
| chr13_-_103053946 | 1.85 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr10_+_88428370 | 1.85 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr8_+_30496078 | 1.84 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr2_-_175711924 | 1.83 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr12_-_99548524 | 1.82 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_69915363 | 1.80 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr6_-_127840021 | 1.80 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr18_+_43307294 | 1.79 |
ENST00000590246.1
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr18_+_8717369 | 1.79 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr12_+_20848486 | 1.78 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr3_+_69915385 | 1.75 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_16738451 | 1.75 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr18_-_70211691 | 1.75 |
ENST00000269503.4
|
CBLN2
|
cerebellin 2 precursor |
| chr15_+_42697018 | 1.73 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr1_+_233765353 | 1.73 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr1_+_66999268 | 1.68 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr22_-_36013368 | 1.64 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr10_-_103603523 | 1.64 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr20_+_20033158 | 1.63 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr13_+_76378407 | 1.63 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr8_+_65285851 | 1.62 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr5_+_59783540 | 1.62 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr11_-_83436446 | 1.60 |
ENST00000529399.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_-_193155729 | 1.60 |
ENST00000367434.4
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr12_+_51785057 | 1.57 |
ENST00000535225.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr2_+_124782857 | 1.57 |
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5 |
| chr17_+_12692774 | 1.56 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr12_+_56477093 | 1.55 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr6_+_69942915 | 1.54 |
ENST00000604969.1
ENST00000603207.1 |
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr15_+_43803143 | 1.53 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr10_-_61122220 | 1.52 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr5_-_36301984 | 1.51 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr3_-_33686743 | 1.50 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_172950227 | 1.50 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr2_+_187454749 | 1.49 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr3_-_73673991 | 1.48 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr6_+_54173227 | 1.47 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr6_-_127840453 | 1.47 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr8_+_54764346 | 1.47 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chrX_+_105412290 | 1.45 |
ENST00000357175.2
ENST00000337685.2 |
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr6_-_29648887 | 1.44 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr10_-_14372870 | 1.44 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr11_-_85338311 | 1.43 |
ENST00000376104.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_-_153599426 | 1.43 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr11_-_41481135 | 1.43 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chrX_+_65384052 | 1.43 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr12_+_48516357 | 1.42 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chrX_-_102565858 | 1.42 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr17_-_41977964 | 1.42 |
ENST00000377184.3
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr1_-_153599732 | 1.40 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr5_+_140734570 | 1.40 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr18_+_55712915 | 1.40 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_+_62462541 | 1.39 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr3_-_165555200 | 1.39 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr19_-_40724246 | 1.38 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr6_+_74405501 | 1.37 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr12_-_10321754 | 1.35 |
ENST00000539518.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_-_22089845 | 1.35 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_-_34420259 | 1.34 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr18_+_32173276 | 1.33 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr6_-_127840048 | 1.33 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_2066252 | 1.33 |
ENST00000466352.1
|
PRKCZ
|
protein kinase C, zeta |
| chr12_+_53443963 | 1.32 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr4_+_71587669 | 1.32 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_173600565 | 1.32 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_48516463 | 1.31 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr12_-_71182695 | 1.30 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr20_-_9819479 | 1.28 |
ENST00000378423.1
ENST00000353224.5 |
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr4_-_152147579 | 1.28 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr18_+_55816546 | 1.27 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_-_224467002 | 1.27 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chrX_+_65384182 | 1.27 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr4_+_159727222 | 1.27 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_-_142888573 | 1.26 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chrX_+_92929192 | 1.25 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr20_-_45980621 | 1.25 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr4_+_71588372 | 1.23 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr14_+_37641012 | 1.22 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr20_-_9819674 | 1.22 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr20_-_56286479 | 1.22 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr15_-_83240553 | 1.22 |
ENST00000423133.2
ENST00000398591.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_+_65266507 | 1.21 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr5_+_140868717 | 1.21 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr3_+_186501979 | 1.21 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr3_-_46904946 | 1.20 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr15_-_77197781 | 1.20 |
ENST00000564590.1
|
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr11_+_111789580 | 1.20 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr8_-_22089533 | 1.19 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr2_-_220252603 | 1.19 |
ENST00000322176.7
ENST00000273075.4 |
DNPEP
|
aspartyl aminopeptidase |
| chr1_-_205904950 | 1.18 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chrX_-_117119243 | 1.17 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr15_-_83240507 | 1.16 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_+_148593425 | 1.16 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_+_20878932 | 1.15 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr2_+_173600514 | 1.15 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_78359999 | 1.14 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr1_+_166958497 | 1.14 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chrX_-_48858667 | 1.13 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr17_-_9683238 | 1.12 |
ENST00000571771.1
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr2_+_162272605 | 1.12 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr3_-_46904918 | 1.12 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr3_+_120626919 | 1.12 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chrX_-_15402498 | 1.12 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr6_-_76203454 | 1.11 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_-_248845629 | 1.11 |
ENST00000342623.3
|
OR14I1
|
olfactory receptor, family 14, subfamily I, member 1 |
| chrX_+_107683096 | 1.11 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr3_+_148508845 | 1.11 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr3_+_75955817 | 1.10 |
ENST00000487694.3
ENST00000602589.1 |
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr9_+_103340354 | 1.10 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr3_-_196242233 | 1.09 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr4_-_186733119 | 1.09 |
ENST00000419063.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_197664366 | 1.08 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr16_+_66878282 | 1.07 |
ENST00000338437.2
|
CA7
|
carbonic anhydrase VII |
| chr15_-_77197620 | 1.06 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr13_-_48575443 | 1.06 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr3_+_173113898 | 1.05 |
ENST00000423427.1
|
NLGN1
|
neuroligin 1 |
| chr13_-_53422640 | 1.05 |
ENST00000338862.4
ENST00000377942.3 |
PCDH8
|
protocadherin 8 |
| chr18_+_7754957 | 1.04 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr2_-_220252530 | 1.04 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr1_+_12042015 | 1.04 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr3_+_190917023 | 1.04 |
ENST00000445281.1
|
OSTN
|
osteocrin |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 1.4 | 4.3 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 1.4 | 4.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.1 | 4.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.1 | 7.7 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.8 | 5.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.8 | 4.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.8 | 3.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.8 | 12.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.7 | 2.9 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.6 | 2.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 3.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.5 | 2.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.5 | 1.5 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.5 | 5.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 6.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.5 | 1.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.5 | 5.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 6.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 2.7 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.4 | 3.1 | GO:0035900 | isocitrate metabolic process(GO:0006102) response to isolation stress(GO:0035900) |
| 0.4 | 2.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 4.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.5 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 | 2.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 2.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 1.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.3 | 4.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 3.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 6.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 2.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.3 | 3.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 1.1 | GO:0098923 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.3 | 1.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 3.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.9 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 0.7 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 1.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 | 3.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.0 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.2 | 1.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 1.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.6 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.2 | 3.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 1.0 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 0.8 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 1.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 1.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.1 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 2.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 2.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 2.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 1.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 2.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.6 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.2 | 0.5 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.2 | 7.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.8 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.7 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.5 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 1.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 1.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 1.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.1 | GO:0097102 | endothelial tip cell fate specification(GO:0097102) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 3.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 1.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 6.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.5 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) operant conditioning(GO:0035106) sperm ejaculation(GO:0042713) |
| 0.1 | 0.9 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.7 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 1.5 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 4.5 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 2.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 1.0 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.6 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 2.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 3.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.0 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.4 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 3.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 1.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:0010482 | ectoderm and mesoderm interaction(GO:0007499) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 3.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.8 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 2.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 2.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 3.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.9 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 1.1 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 1.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 1.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 1.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:1901490 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.7 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 2.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 1.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 2.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 1.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 1.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.8 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.7 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 2.8 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.6 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.7 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:1904776 | protein localization to cell cortex(GO:0072697) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.5 | 5.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 4.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 1.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.5 | 2.0 | GO:0030849 | autosome(GO:0030849) |
| 0.5 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 4.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 2.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 4.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 2.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 6.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 4.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 18.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 5.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 1.7 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 4.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 7.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 3.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 3.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 6.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 1.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 7.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 23.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 4.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 9.7 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 4.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 4.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 8.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.3 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 3.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 3.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 1.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 3.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 1.0 | 3.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 1.0 | 4.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 8.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 4.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 6.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 6.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 2.9 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.5 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 13.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 2.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.5 | 3.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.4 | 1.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 3.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.4 | 3.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 2.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 1.0 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.3 | 8.4 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.3 | 1.8 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 2.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 4.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 1.4 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.3 | 0.8 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.3 | 1.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.3 | 2.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.2 | 2.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 0.6 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.2 | 12.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 0.7 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 5.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 2.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 3.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 4.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.7 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 3.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 4.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 3.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 2.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 1.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 3.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 3.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 1.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 3.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 1.6 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 2.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.6 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 2.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 7.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 3.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 8.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 4.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 6.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 4.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 5.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 5.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 5.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |