Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
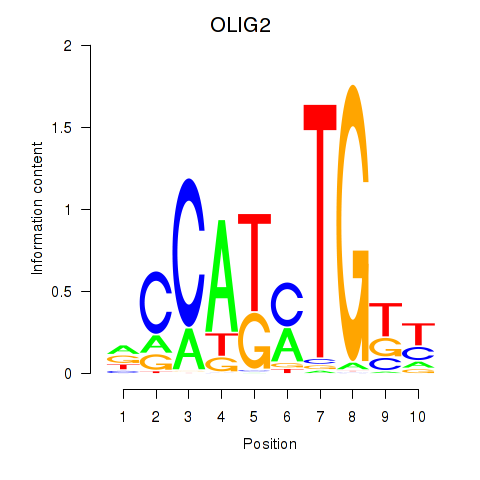
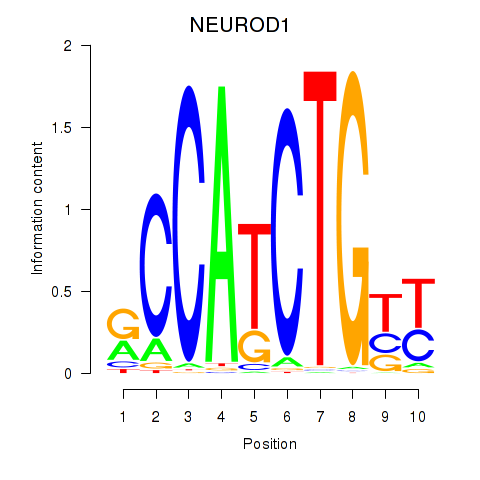
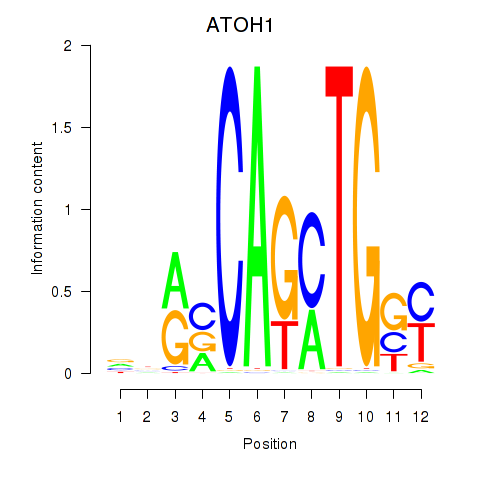
Results for OLIG2_NEUROD1_ATOH1
Z-value: 1.73
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | oligodendrocyte transcription factor 2 |
|
NEUROD1
|
ENSG00000162992.3 | neuronal differentiation 1 |
|
ATOH1
|
ENSG00000172238.3 | atonal bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATOH1 | hg19_v2_chr4_+_94750014_94750042 | 0.29 | 1.1e-01 | Click! |
| OLIG2 | hg19_v2_chr21_+_34398153_34398250 | 0.25 | 1.6e-01 | Click! |
| NEUROD1 | hg19_v2_chr2_-_182545603_182545603 | -0.20 | 2.7e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_195310802 | 7.73 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr4_+_114214125 | 4.23 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr15_-_94614049 | 4.08 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr19_-_45826125 | 3.70 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr5_+_71475449 | 3.55 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr6_+_53948328 | 3.43 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr1_-_171621815 | 3.40 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr19_-_49944806 | 3.17 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr2_-_152590946 | 3.04 |
ENST00000172853.10
|
NEB
|
nebulin |
| chr6_+_53948221 | 2.68 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr7_+_15728003 | 2.65 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chrX_+_135279179 | 2.57 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr4_+_113970772 | 2.56 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr10_-_105437909 | 2.53 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr19_-_35626104 | 2.38 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr5_-_59481406 | 2.36 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chrX_+_135278908 | 2.27 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr2_-_241759622 | 2.27 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr1_+_159141397 | 2.24 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr19_-_6501778 | 2.20 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr19_-_38743878 | 2.18 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr10_-_97321112 | 2.17 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chrX_-_33357558 | 2.14 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr5_+_173472607 | 2.13 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr6_-_119031228 | 2.10 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr10_-_97321165 | 2.04 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr10_-_61513201 | 2.01 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr3_+_183948161 | 1.98 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr9_+_113431029 | 1.95 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr6_+_155443048 | 1.95 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chrX_+_100474711 | 1.89 |
ENST00000402866.1
|
DRP2
|
dystrophin related protein 2 |
| chr19_-_46285736 | 1.87 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chr8_-_41522779 | 1.85 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr2_-_163100045 | 1.83 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr1_-_165414414 | 1.82 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr19_-_46285646 | 1.82 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr17_-_15168624 | 1.81 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chrX_+_100474906 | 1.79 |
ENST00000541709.1
|
DRP2
|
dystrophin related protein 2 |
| chr3_+_35722424 | 1.78 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chrX_+_100474763 | 1.73 |
ENST00000395209.3
|
DRP2
|
dystrophin related protein 2 |
| chr1_+_19967014 | 1.71 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr2_+_233404429 | 1.70 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr16_+_6533729 | 1.66 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr10_-_61513146 | 1.64 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr1_+_226736446 | 1.62 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr16_+_30383613 | 1.62 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr20_-_52645231 | 1.54 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr17_+_33474860 | 1.52 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr6_-_125766306 | 1.50 |
ENST00000430672.1
|
RP11-138M12.1
|
RP11-138M12.1 |
| chr14_-_75078725 | 1.50 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr5_-_180632147 | 1.47 |
ENST00000274773.7
|
TRIM7
|
tripartite motif containing 7 |
| chr9_+_113431059 | 1.47 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr3_-_18480173 | 1.46 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr11_-_41481135 | 1.44 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr10_-_13043697 | 1.43 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr1_-_154842741 | 1.43 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr19_+_46806856 | 1.42 |
ENST00000300862.3
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr7_-_44105158 | 1.41 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr11_-_1587166 | 1.40 |
ENST00000331588.4
|
DUSP8
|
dual specificity phosphatase 8 |
| chr3_-_18480260 | 1.40 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr9_+_70971815 | 1.40 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr17_+_33474826 | 1.39 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr5_-_180632293 | 1.37 |
ENST00000334421.5
|
TRIM7
|
tripartite motif containing 7 |
| chr12_+_54393880 | 1.36 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr2_-_183387064 | 1.34 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_154335300 | 1.34 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr13_-_36705425 | 1.34 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr2_-_152830441 | 1.34 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_-_177190364 | 1.32 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr12_+_101988627 | 1.32 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr17_-_41132088 | 1.31 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_42727011 | 1.31 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr11_-_57417405 | 1.29 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr3_+_99833755 | 1.28 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr12_+_79371565 | 1.28 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr15_-_40633101 | 1.28 |
ENST00000559313.1
|
C15orf52
|
chromosome 15 open reading frame 52 |
| chr3_+_85008089 | 1.28 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr12_-_57634475 | 1.25 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr4_-_108204846 | 1.23 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr16_+_56385290 | 1.22 |
ENST00000564727.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr3_-_99833333 | 1.19 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr4_+_169633310 | 1.19 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_101988774 | 1.19 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr3_+_159570722 | 1.18 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_-_30144432 | 1.15 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr9_+_19408919 | 1.14 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr16_+_7560114 | 1.14 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_+_113391696 | 1.14 |
ENST00000512927.1
|
RP11-371M22.1
|
RP11-371M22.1 |
| chr9_-_35689900 | 1.13 |
ENST00000378300.5
ENST00000329305.2 ENST00000360958.2 |
TPM2
|
tropomyosin 2 (beta) |
| chr8_-_41522719 | 1.12 |
ENST00000335651.6
|
ANK1
|
ankyrin 1, erythrocytic |
| chr14_+_93389425 | 1.12 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr8_+_22411931 | 1.08 |
ENST00000523402.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_-_111174054 | 1.08 |
ENST00000369770.3
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr7_-_99869799 | 1.07 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr16_+_30386098 | 1.06 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr19_+_1452188 | 1.06 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr6_-_30080863 | 1.05 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr6_+_53976235 | 1.02 |
ENST00000502396.1
ENST00000358276.5 |
MLIP
|
muscular LMNA-interacting protein |
| chr2_+_220325441 | 1.01 |
ENST00000396688.1
|
SPEG
|
SPEG complex locus |
| chr20_+_30407105 | 1.01 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr20_+_30407151 | 1.00 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chrX_-_99665262 | 1.00 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr1_-_243326612 | 1.00 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr3_+_69788576 | 0.98 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_7513904 | 0.98 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr6_+_53976211 | 0.97 |
ENST00000503951.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr17_-_3301704 | 0.97 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr1_-_26394114 | 0.96 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr3_-_52868931 | 0.96 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr9_+_140033862 | 0.95 |
ENST00000350902.5
ENST00000371550.4 ENST00000371546.4 ENST00000371555.4 ENST00000371553.3 ENST00000371559.4 ENST00000371560.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr7_+_123295861 | 0.94 |
ENST00000458573.2
ENST00000456238.2 |
LMOD2
|
leiomodin 2 (cardiac) |
| chr10_-_72141330 | 0.94 |
ENST00000395011.1
ENST00000395010.1 |
LRRC20
|
leucine rich repeat containing 20 |
| chr17_+_65040678 | 0.94 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr10_+_18429671 | 0.94 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr8_+_86351056 | 0.93 |
ENST00000285381.2
|
CA3
|
carbonic anhydrase III, muscle specific |
| chr1_+_201159914 | 0.92 |
ENST00000335211.4
ENST00000451870.2 ENST00000295591.8 |
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr19_-_18902106 | 0.92 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr11_+_46299199 | 0.91 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr6_-_30080876 | 0.91 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr5_+_102201687 | 0.91 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_+_43809797 | 0.90 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr5_+_102201430 | 0.90 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_31683117 | 0.89 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr5_+_102201722 | 0.88 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_102201509 | 0.88 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_79792909 | 0.87 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr2_+_29001711 | 0.87 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr6_-_42690312 | 0.86 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr5_+_173472744 | 0.86 |
ENST00000521585.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr12_+_53491220 | 0.85 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr3_+_148508845 | 0.82 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr8_-_22014255 | 0.82 |
ENST00000424267.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr17_-_9725388 | 0.82 |
ENST00000399363.4
|
RP11-477N12.3
|
Putative germ cell-specific gene 1-like protein 2 |
| chr2_+_79740118 | 0.82 |
ENST00000496558.1
ENST00000451966.1 |
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr17_+_7341586 | 0.82 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr15_-_42749711 | 0.82 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr14_-_75079294 | 0.81 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr14_-_65289812 | 0.80 |
ENST00000389720.3
ENST00000389721.5 ENST00000389722.3 |
SPTB
|
spectrin, beta, erythrocytic |
| chr6_+_53976285 | 0.80 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr16_+_56623433 | 0.80 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr17_+_48243352 | 0.79 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr3_-_52869205 | 0.79 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr7_+_75931861 | 0.79 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr12_-_130529501 | 0.79 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr10_+_18429606 | 0.78 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr9_+_131174024 | 0.77 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr14_-_75079026 | 0.77 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr22_+_31488433 | 0.77 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr11_+_133902667 | 0.76 |
ENST00000533091.1
ENST00000527712.1 |
RP11-713P17.3
|
RP11-713P17.3 |
| chr3_+_111717511 | 0.76 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr19_-_47164386 | 0.76 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr22_+_42372970 | 0.76 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr12_+_48152774 | 0.75 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr1_+_86934526 | 0.75 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr10_+_25463951 | 0.74 |
ENST00000376351.3
|
GPR158
|
G protein-coupled receptor 158 |
| chr19_+_35596873 | 0.74 |
ENST00000313865.6
|
AC020907.1
|
Uncharacterized protein |
| chr1_-_201081579 | 0.74 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr2_-_183387283 | 0.73 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_29555683 | 0.72 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr3_+_132843652 | 0.71 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr5_-_159766528 | 0.71 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr12_-_48152428 | 0.71 |
ENST00000449771.2
ENST00000395358.3 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_-_62066769 | 0.71 |
ENST00000577329.1
|
CTC-264K15.6
|
CTC-264K15.6 |
| chr14_-_21109850 | 0.71 |
ENST00000320704.3
|
OR6S1
|
olfactory receptor, family 6, subfamily S, member 1 |
| chr1_-_242162375 | 0.70 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr3_+_54157480 | 0.70 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr16_+_6533380 | 0.70 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_240969906 | 0.70 |
ENST00000402971.2
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2 |
| chr19_-_49222956 | 0.70 |
ENST00000599703.1
ENST00000318083.6 ENST00000419611.1 ENST00000377367.3 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr17_-_41738931 | 0.70 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr2_-_183387430 | 0.70 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_99470558 | 0.70 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr19_-_47137942 | 0.69 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chrX_+_70521584 | 0.69 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr19_-_11591848 | 0.68 |
ENST00000359227.3
|
ELAVL3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_-_81916501 | 0.68 |
ENST00000555001.1
|
RP11-299L17.3
|
RP11-299L17.3 |
| chr5_+_156887027 | 0.68 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr15_-_78526855 | 0.67 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr7_-_74867509 | 0.67 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr1_+_46379254 | 0.67 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr19_+_35521699 | 0.67 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr14_+_69726656 | 0.66 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr7_-_30029574 | 0.66 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr19_+_18846389 | 0.66 |
ENST00000594658.1
|
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr3_+_8775466 | 0.66 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr7_-_30029328 | 0.65 |
ENST00000425819.2
ENST00000409570.1 |
SCRN1
|
secernin 1 |
| chr11_-_133402410 | 0.65 |
ENST00000524381.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chrX_-_13835398 | 0.65 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr3_-_52002194 | 0.65 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr15_+_74466744 | 0.64 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr8_-_22014339 | 0.64 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr19_-_2256405 | 0.63 |
ENST00000300961.6
|
JSRP1
|
junctional sarcoplasmic reticulum protein 1 |
| chr16_+_67280799 | 0.63 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr17_-_67057203 | 0.63 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr4_-_186733363 | 0.63 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_+_7231123 | 0.63 |
ENST00000383467.2
|
LRRC30
|
leucine rich repeat containing 30 |
| chr17_-_67057114 | 0.62 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr3_+_155755482 | 0.62 |
ENST00000472028.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_-_32173579 | 0.62 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr8_+_39972170 | 0.62 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.4 | 6.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.1 | 3.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.9 | 3.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.6 | 1.8 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.6 | 3.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.6 | 3.6 | GO:0018032 | protein amidation(GO:0018032) |
| 0.4 | 6.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.4 | 1.2 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 1.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 3.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.4 | 1.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.4 | 1.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 1.7 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 1.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.3 | 3.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 1.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 1.7 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.3 | 2.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 0.9 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.3 | 1.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.3 | 1.9 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.3 | 0.8 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.3 | 1.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 1.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.3 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.2 | 0.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.2 | 1.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 0.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.2 | 1.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 0.5 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 3.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.2 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 3.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 2.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 3.4 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 3.0 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 1.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.4 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.3 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 3.1 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 2.8 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.6 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.5 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 2.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.7 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 1.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.2 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.1 | 5.3 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.1 | 2.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.2 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 1.1 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 3.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 1.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 1.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.6 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.6 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 1.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 3.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.5 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 9.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.8 | GO:0043271 | negative regulation of ion transport(GO:0043271) |
| 0.0 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.5 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.6 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.2 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 1.0 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.9 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.4 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.6 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.7 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.5 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.2 | GO:0001654 | eye development(GO:0001654) |
| 0.0 | 7.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.6 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.5 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 1.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.6 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.6 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 1.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0010628 | positive regulation of gene expression(GO:0010628) |
| 0.0 | 0.1 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0048284 | organelle fusion(GO:0048284) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 1.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 2.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 1.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 1.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 1.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.3 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 4.4 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.2 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 2.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 14.8 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.8 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 5.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 4.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 2.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 2.9 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 2.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.4 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 4.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 11.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 9.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 6.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 8.4 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 3.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 4.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 3.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 1.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.6 | 1.7 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.5 | 1.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 2.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 1.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.4 | 3.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 3.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.4 | 1.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 3.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.8 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 1.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.9 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.2 | 0.9 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.2 | 0.9 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 3.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 10.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 3.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 2.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 11.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 7.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.4 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.6 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.6 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 6.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 6.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 6.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 9.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 1.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 5.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 4.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 6.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 5.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 3.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 3.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 7.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |