Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
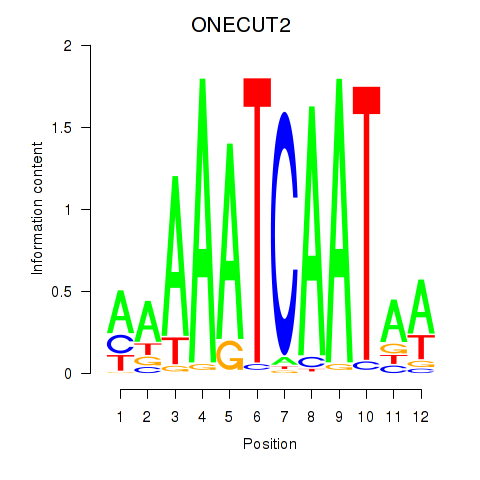
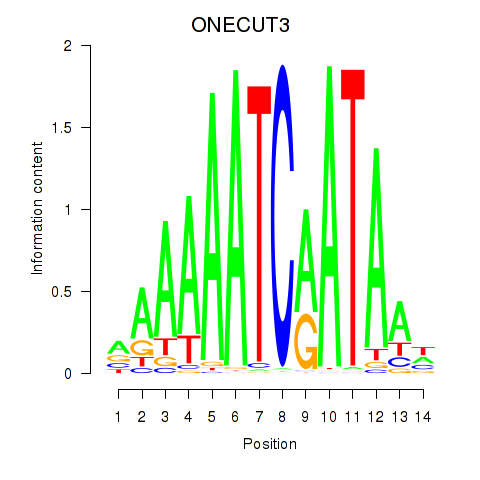
Results for ONECUT2_ONECUT3
Z-value: 1.61
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | 0.22 | 2.4e-01 | Click! |
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | 0.18 | 3.1e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63428752 | 7.28 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr14_-_24047965 | 5.51 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr10_+_95517616 | 4.43 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr10_+_95517660 | 4.28 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr10_+_95517566 | 4.00 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr12_-_62586543 | 3.53 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr11_+_134144139 | 3.50 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr8_+_77316233 | 3.39 |
ENST00000603284.1
ENST00000603837.1 |
RP11-706J10.2
RP11-706J10.3
|
RP11-706J10.2 long intergenic non-protein coding RNA 1109 |
| chr2_-_192016316 | 3.27 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr3_-_187388173 | 3.23 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr2_-_192016276 | 2.96 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr16_+_72459838 | 2.93 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr3_+_63428982 | 2.76 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr1_-_116383322 | 2.57 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr1_+_198608292 | 2.50 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_116383738 | 2.46 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr11_-_35440796 | 2.38 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr20_-_1974692 | 2.36 |
ENST00000217305.2
ENST00000539905.1 |
PDYN
|
prodynorphin |
| chr5_+_126984710 | 2.30 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr14_+_77607026 | 2.29 |
ENST00000600936.1
|
AC007375.1
|
Uncharacterized protein; cDNA FLJ43210 fis, clone FEBRA2020582 |
| chr1_+_198607801 | 2.23 |
ENST00000367379.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_30009542 | 2.22 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr1_+_198608146 | 2.16 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr5_-_156772729 | 2.12 |
ENST00000312349.4
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr2_+_58655520 | 2.07 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_-_220174166 | 2.03 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr5_-_160279207 | 2.02 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr13_-_36429763 | 1.98 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr17_+_61151306 | 1.97 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_-_30008849 | 1.91 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr7_-_143059780 | 1.88 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr1_+_190448095 | 1.81 |
ENST00000424735.1
|
RP11-547I7.2
|
RP11-547I7.2 |
| chr7_-_143059845 | 1.77 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr2_+_166095898 | 1.75 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr13_-_84456527 | 1.74 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr18_-_70211691 | 1.68 |
ENST00000269503.4
|
CBLN2
|
cerebellin 2 precursor |
| chr7_-_107968921 | 1.64 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_111150048 | 1.63 |
ENST00000485317.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr11_-_35440579 | 1.61 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_104909435 | 1.61 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr7_-_107968999 | 1.60 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_-_100239132 | 1.56 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr5_-_156772568 | 1.55 |
ENST00000520782.1
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr14_+_62585332 | 1.54 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr5_-_82969405 | 1.51 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr6_+_15401075 | 1.49 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_-_37764128 | 1.49 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr10_+_68685764 | 1.48 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr17_+_44701402 | 1.44 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_-_27924209 | 1.39 |
ENST00000381273.3
|
MANSC4
|
MANSC domain containing 4 |
| chr2_-_50201327 | 1.28 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr2_+_215275771 | 1.25 |
ENST00000312504.5
ENST00000427124.1 |
VWC2L
|
von Willebrand factor C domain containing protein 2-like |
| chr7_-_121944491 | 1.22 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr1_-_26233423 | 1.22 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr1_-_243326612 | 1.17 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr7_+_43152212 | 1.16 |
ENST00000453890.1
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_129906660 | 1.11 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr14_+_22265444 | 1.10 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr5_+_161277603 | 1.10 |
ENST00000519621.1
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_-_20985632 | 1.09 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr4_-_102267953 | 1.08 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_+_37475109 | 1.06 |
ENST00000570443.2
|
RP1-153P14.8
|
RP1-153P14.8 |
| chr9_-_28670283 | 1.06 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr8_+_104831440 | 1.04 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_46930171 | 1.04 |
ENST00000593391.1
|
AC109583.1
|
Uncharacterized protein |
| chr19_+_3880581 | 1.03 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr17_-_2304365 | 1.03 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr2_+_105050794 | 1.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chrX_+_123480194 | 1.01 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr6_-_116479897 | 0.99 |
ENST00000418500.1
|
COL10A1
|
collagen, type X, alpha 1 |
| chr4_-_70826725 | 0.99 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr16_+_25228242 | 0.95 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr14_+_77425972 | 0.95 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr2_-_55646412 | 0.89 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr10_+_70980051 | 0.85 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr11_-_107729287 | 0.84 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_+_77230499 | 0.83 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr12_-_68797580 | 0.82 |
ENST00000539404.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr1_+_117297007 | 0.81 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr11_-_107729504 | 0.80 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_+_20848377 | 0.80 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr2_-_163695128 | 0.79 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr3_-_155394099 | 0.78 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr7_+_39125365 | 0.77 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr12_+_20848282 | 0.77 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr2_+_157330081 | 0.76 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr7_+_43152191 | 0.76 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_-_130745403 | 0.75 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr3_-_108836945 | 0.74 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr14_+_22782867 | 0.73 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40 |
| chr1_+_206730484 | 0.73 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr11_+_60467047 | 0.71 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr12_+_4385230 | 0.71 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr6_+_126221034 | 0.70 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_-_186080012 | 0.70 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr16_-_28192360 | 0.69 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr14_+_88490894 | 0.68 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr15_+_54901540 | 0.68 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr3_-_155394152 | 0.67 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr13_-_46952645 | 0.67 |
ENST00000439642.1
|
KIAA0226L
|
KIAA0226-like |
| chr4_+_156824840 | 0.66 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr2_-_208030886 | 0.64 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chrX_+_47093171 | 0.64 |
ENST00000377078.2
|
USP11
|
ubiquitin specific peptidase 11 |
| chr3_-_194119083 | 0.64 |
ENST00000401815.1
|
GP5
|
glycoprotein V (platelet) |
| chr10_-_62332357 | 0.63 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_205538165 | 0.62 |
ENST00000536357.1
|
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr12_+_9144626 | 0.62 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr19_+_7598890 | 0.61 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr17_-_79876010 | 0.61 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr8_+_31497271 | 0.59 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr7_-_28220354 | 0.59 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr3_+_121774202 | 0.58 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr4_-_69434245 | 0.57 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr12_-_10601963 | 0.56 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_+_4510109 | 0.55 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chr2_-_214017151 | 0.55 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_-_102268628 | 0.55 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_-_163695238 | 0.54 |
ENST00000328032.4
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr4_+_174818390 | 0.54 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr11_-_107729887 | 0.54 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chrX_-_119693745 | 0.53 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr10_-_121296045 | 0.53 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr7_-_151330218 | 0.52 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr20_+_58571419 | 0.51 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr12_+_64846129 | 0.51 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr9_+_74729511 | 0.50 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr14_+_23004404 | 0.49 |
ENST00000390528.1
|
TRAJ9
|
T cell receptor alpha joining 9 |
| chr17_+_7461613 | 0.48 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_-_102268708 | 0.48 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_108863651 | 0.47 |
ENST00000329106.2
ENST00000376700.1 |
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3 |
| chr11_-_47736896 | 0.47 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr12_+_100594557 | 0.47 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr6_+_105404899 | 0.47 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr9_+_95726243 | 0.46 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr7_-_112727774 | 0.46 |
ENST00000297146.3
ENST00000501255.2 |
GPR85
|
G protein-coupled receptor 85 |
| chr17_-_4167142 | 0.45 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr21_-_15918618 | 0.45 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr16_-_75467318 | 0.44 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr11_-_60623437 | 0.44 |
ENST00000332539.4
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr7_+_128784712 | 0.43 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr19_+_7599128 | 0.43 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr12_-_122018114 | 0.43 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_+_110655050 | 0.43 |
ENST00000334179.3
|
UBL4B
|
ubiquitin-like 4B |
| chr1_-_182921119 | 0.42 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr3_-_71632894 | 0.42 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr9_-_117420052 | 0.41 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr7_-_23510086 | 0.41 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr12_+_41831485 | 0.41 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr16_+_73266661 | 0.41 |
ENST00000561802.1
|
AC140912.1
|
AC140912.1 |
| chr2_+_168725458 | 0.40 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr19_+_35940486 | 0.39 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr1_+_114473350 | 0.38 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr12_+_64845864 | 0.38 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr17_+_7461849 | 0.37 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr7_+_129007964 | 0.37 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr14_+_29241910 | 0.37 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr4_-_102268484 | 0.36 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chrX_+_10124977 | 0.36 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr2_+_58655461 | 0.36 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr6_+_56954919 | 0.35 |
ENST00000508603.1
ENST00000491832.2 ENST00000370710.6 |
ZNF451
|
zinc finger protein 451 |
| chr2_+_102413726 | 0.34 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr18_+_11752783 | 0.34 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr2_+_7017796 | 0.33 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr6_-_113754604 | 0.33 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr12_-_122017542 | 0.33 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_+_175839551 | 0.33 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr2_-_32490859 | 0.32 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr7_-_78400598 | 0.32 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_175839506 | 0.32 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr14_+_23012122 | 0.32 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr16_-_30621663 | 0.31 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr1_+_223354486 | 0.31 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr6_+_27925019 | 0.31 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr19_+_37997812 | 0.30 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr17_+_31318886 | 0.29 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr15_+_48051920 | 0.29 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chrX_-_133792480 | 0.28 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr5_+_162887556 | 0.28 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr17_+_17685422 | 0.27 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr5_-_121659052 | 0.27 |
ENST00000512105.1
|
CTD-2544H17.1
|
CTD-2544H17.1 |
| chr16_-_75467274 | 0.26 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr15_+_36338242 | 0.26 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr6_+_158733692 | 0.25 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr9_+_33629119 | 0.24 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr5_+_180794269 | 0.24 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor, family 4, subfamily F, member 3 |
| chr8_-_117043 | 0.24 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr7_+_133615169 | 0.24 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chrX_-_16887963 | 0.24 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr1_+_248201474 | 0.24 |
ENST00000366479.2
|
OR2L2
|
olfactory receptor, family 2, subfamily L, member 2 |
| chr1_-_146068184 | 0.24 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr17_+_7461781 | 0.23 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_+_4699244 | 0.22 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_+_367640 | 0.22 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr18_+_29671812 | 0.22 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr9_-_27529726 | 0.22 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr4_+_71587669 | 0.22 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_71588372 | 0.22 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_-_162873821 | 0.22 |
ENST00000599797.1
|
AC112205.1
|
Uncharacterized protein |
| chr12_+_32832134 | 0.21 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr12_+_48866448 | 0.21 |
ENST00000266594.1
|
ANP32D
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member D |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.6 | 3.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.6 | 2.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 5.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 4.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 1.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 1.6 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 1.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.2 | 1.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 0.7 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 | 0.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 3.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.6 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.2 | 0.7 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.8 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.2 | 0.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 0.6 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.4 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 4.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.7 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.8 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.5 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 4.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 12.7 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 1.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 5.2 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 2.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.2 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.7 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.0 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.7 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 1.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.8 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.7 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 10.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 3.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 4.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 1.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 4.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 16.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.4 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 2.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 3.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 0.8 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 2.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 6.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.4 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 3.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 1.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.8 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 3.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 2.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.2 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 8.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 5.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 3.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |