Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for OSR1_OSR2
Z-value: 1.58
Transcription factors associated with OSR1_OSR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
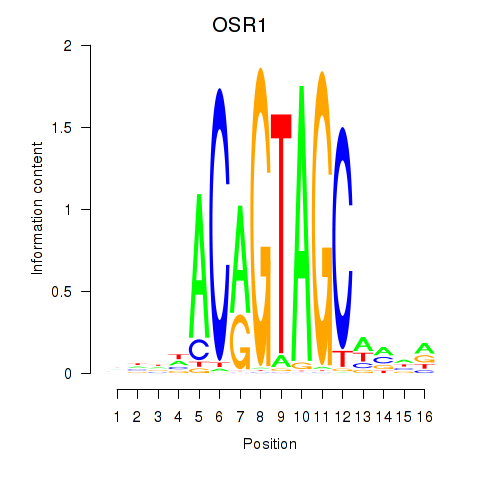
OSR1
|
ENSG00000143867.5 | odd-skipped related transcription factor 1 |
|
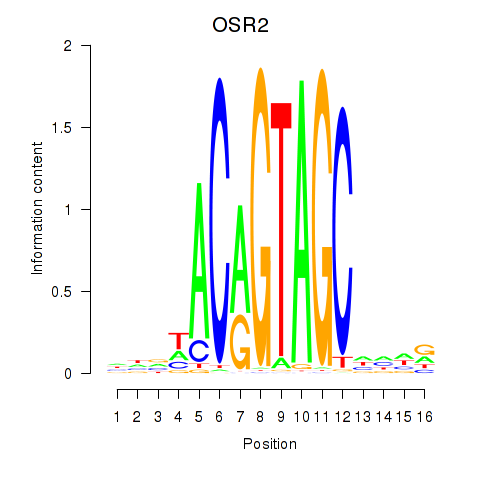
OSR2
|
ENSG00000164920.5 | odd-skipped related transciption factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OSR2 | hg19_v2_chr8_+_99956759_99956887 | -0.26 | 1.5e-01 | Click! |
| OSR1 | hg19_v2_chr2_-_19558373_19558414 | -0.13 | 4.9e-01 | Click! |
Activity profile of OSR1_OSR2 motif
Sorted Z-values of OSR1_OSR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_70361615 | 6.23 |
ENST00000305107.6
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4 |
| chr4_-_70361579 | 6.16 |
ENST00000512583.1
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4 |
| chr16_-_31439735 | 5.78 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr4_+_169013666 | 4.51 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr5_-_147211226 | 4.41 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr4_-_70080449 | 4.30 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr1_-_151342180 | 3.82 |
ENST00000424475.1
|
SELENBP1
|
selenium binding protein 1 |
| chr2_+_234959323 | 3.67 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr3_+_149191723 | 2.93 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr3_+_108855558 | 2.88 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr2_+_234959376 | 2.85 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr2_-_88125471 | 2.78 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr3_+_149192475 | 2.76 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr7_+_134212312 | 2.75 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr2_-_105030466 | 2.67 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr4_+_69681710 | 2.65 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr19_-_45826125 | 2.64 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr7_-_95225768 | 2.59 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr14_+_21152259 | 2.51 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr1_-_237167718 | 2.50 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr16_-_56701933 | 2.43 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr2_+_234580525 | 2.40 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_+_185734773 | 2.36 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr2_+_234580499 | 2.33 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_-_42887494 | 2.20 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_186696425 | 1.98 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_186696515 | 1.89 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_196857144 | 1.89 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr3_-_50360192 | 1.85 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr1_+_12042015 | 1.85 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr14_+_21152706 | 1.77 |
ENST00000397995.2
ENST00000304704.4 ENST00000553909.1 |
RNASE4
AL163636.6
|
ribonuclease, RNase A family, 4 Homo sapiens ribonuclease, RNase A family, 4 (RNASE4), transcript variant 4, mRNA. |
| chr1_+_196743943 | 1.76 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr4_-_186696561 | 1.68 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_76248538 | 1.67 |
ENST00000274368.4
|
CRHBP
|
corticotropin releasing hormone binding protein |
| chr1_+_196743912 | 1.67 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr3_+_193853927 | 1.63 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr15_+_58724184 | 1.57 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr8_+_66619277 | 1.48 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr4_-_186697044 | 1.46 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185733394 | 1.40 |
ENST00000504342.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr12_+_109273806 | 1.38 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr15_+_63188009 | 1.30 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_-_207143802 | 1.28 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr6_+_152011628 | 1.28 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr4_-_186696636 | 1.21 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_36994210 | 1.20 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr10_-_94301107 | 1.19 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr12_-_9268819 | 1.18 |
ENST00000404455.2
|
A2M
|
alpha-2-macroglobulin |
| chr12_-_14967095 | 1.15 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr4_+_151503077 | 1.12 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr1_+_47264711 | 1.11 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr17_+_79859985 | 1.11 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr6_-_136788001 | 1.08 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr9_-_95055923 | 1.04 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr9_-_34710066 | 1.03 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr8_-_80942139 | 1.00 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr7_+_16700806 | 0.98 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr2_+_37458904 | 0.98 |
ENST00000416653.1
|
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr5_-_132166579 | 0.93 |
ENST00000378679.3
|
SHROOM1
|
shroom family member 1 |
| chr6_+_10585979 | 0.92 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr4_+_39460659 | 0.90 |
ENST00000513731.1
|
LIAS
|
lipoic acid synthetase |
| chr17_-_191188 | 0.90 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr8_-_80942061 | 0.86 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr2_+_37458776 | 0.84 |
ENST00000002125.4
ENST00000336237.6 ENST00000431821.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr15_+_41523335 | 0.84 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr22_+_31489344 | 0.83 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr8_+_94710789 | 0.83 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr15_+_41523417 | 0.81 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr17_-_46690839 | 0.77 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr3_+_50654821 | 0.76 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr5_+_110073853 | 0.74 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr16_+_71560023 | 0.73 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr14_+_105266933 | 0.73 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr19_+_19144384 | 0.72 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr2_+_37458928 | 0.72 |
ENST00000439218.1
ENST00000432075.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chrX_-_15333775 | 0.71 |
ENST00000480796.1
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr20_-_33999766 | 0.69 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr2_-_183387283 | 0.68 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_43904608 | 0.67 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr8_+_37594130 | 0.67 |
ENST00000518526.1
ENST00000523887.1 ENST00000276461.5 |
ERLIN2
|
ER lipid raft associated 2 |
| chr9_-_95056010 | 0.65 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr1_-_231560790 | 0.65 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr8_-_37594944 | 0.64 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr10_-_36812323 | 0.62 |
ENST00000543053.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr16_-_66968055 | 0.61 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr14_+_105267250 | 0.59 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr10_-_43904235 | 0.58 |
ENST00000356053.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_233749739 | 0.58 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr4_-_8430152 | 0.57 |
ENST00000514423.1
ENST00000503233.1 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr19_+_19144666 | 0.57 |
ENST00000535288.1
ENST00000538663.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr11_+_64808675 | 0.56 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr10_+_78078088 | 0.56 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr15_-_41694640 | 0.55 |
ENST00000558719.1
ENST00000260361.4 ENST00000560978.1 |
NDUFAF1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr4_+_39460689 | 0.54 |
ENST00000381846.1
|
LIAS
|
lipoic acid synthetase |
| chr5_+_135383008 | 0.54 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr19_-_41220957 | 0.53 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr1_-_196577489 | 0.52 |
ENST00000609185.1
ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2
|
potassium channel, subfamily T, member 2 |
| chr22_-_26986045 | 0.51 |
ENST00000442495.1
ENST00000440953.1 ENST00000450022.1 ENST00000338754.4 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr20_-_34542548 | 0.50 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr2_-_190448118 | 0.49 |
ENST00000440626.1
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr16_+_67143880 | 0.49 |
ENST00000219139.3
ENST00000566026.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr8_-_57906362 | 0.46 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr6_+_80341000 | 0.46 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr5_-_125930877 | 0.45 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr4_+_146403912 | 0.45 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr19_-_45457264 | 0.44 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chr4_+_145567173 | 0.44 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr1_-_156722015 | 0.44 |
ENST00000368209.5
|
HDGF
|
hepatoma-derived growth factor |
| chr19_-_13068012 | 0.44 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr1_+_218458625 | 0.44 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr9_-_95055956 | 0.43 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr3_+_130648842 | 0.43 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_156561533 | 0.41 |
ENST00000368234.3
ENST00000368235.3 ENST00000368233.3 |
APOA1BP
|
apolipoprotein A-I binding protein |
| chr7_-_76955563 | 0.39 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chrX_-_134049262 | 0.39 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr15_-_74284613 | 0.39 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr2_-_74735707 | 0.38 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr12_-_58165870 | 0.38 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr19_-_16770915 | 0.37 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr13_-_52703187 | 0.37 |
ENST00000355568.4
|
NEK5
|
NIMA-related kinase 5 |
| chr17_-_79894651 | 0.37 |
ENST00000584848.1
ENST00000577756.1 ENST00000329875.8 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr7_-_43769066 | 0.37 |
ENST00000223336.6
ENST00000310564.6 ENST00000431651.1 ENST00000415798.1 |
COA1
|
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr13_+_97874574 | 0.37 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr1_+_42922173 | 0.36 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr1_-_59043166 | 0.35 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr9_+_33025209 | 0.35 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chrX_-_134049233 | 0.35 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr3_+_179322481 | 0.33 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr7_-_123197733 | 0.33 |
ENST00000470123.1
ENST00000471770.1 |
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chrX_-_15333736 | 0.33 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr8_+_37594103 | 0.33 |
ENST00000397228.2
|
ERLIN2
|
ER lipid raft associated 2 |
| chr5_-_125930929 | 0.33 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr1_-_156722195 | 0.32 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_-_42180940 | 0.31 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr3_+_50211240 | 0.31 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr7_-_43769051 | 0.31 |
ENST00000395880.3
|
COA1
|
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr12_-_10251539 | 0.31 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr14_+_65453432 | 0.30 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr2_-_133104839 | 0.30 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr1_+_201798269 | 0.30 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr14_+_93651296 | 0.30 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chr5_+_69345350 | 0.29 |
ENST00000380741.4
ENST00000380743.4 ENST00000511812.1 ENST00000380742.4 |
SMN2
|
survival of motor neuron 2, centromeric |
| chr7_+_150706010 | 0.29 |
ENST00000475017.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr21_+_17214724 | 0.29 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr8_-_145653885 | 0.29 |
ENST00000531032.1
ENST00000292510.4 ENST00000377348.2 ENST00000530790.1 ENST00000533806.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr17_+_40714092 | 0.29 |
ENST00000420359.1
ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY
|
CoA synthase |
| chr1_+_232940643 | 0.28 |
ENST00000418460.1
|
MAP10
|
microtubule-associated protein 10 |
| chr2_-_37458749 | 0.28 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr1_-_35450897 | 0.28 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr22_-_45559540 | 0.28 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr5_-_36152031 | 0.27 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr5_+_70220768 | 0.27 |
ENST00000351205.4
ENST00000503079.2 ENST00000380707.4 ENST00000514951.1 ENST00000506163.1 |
SMN1
|
survival of motor neuron 1, telomeric |
| chr12_+_58166431 | 0.25 |
ENST00000333012.5
|
METTL21B
|
methyltransferase like 21B |
| chr1_+_38273419 | 0.25 |
ENST00000468084.1
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_-_95467356 | 0.25 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr5_+_36152163 | 0.25 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_+_96037255 | 0.25 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr11_+_43380459 | 0.24 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr20_-_30311703 | 0.24 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr1_+_17906970 | 0.24 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr8_+_37594185 | 0.24 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chr3_-_179322436 | 0.24 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr7_+_73106926 | 0.24 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr13_-_52980263 | 0.23 |
ENST00000258613.4
ENST00000544466.1 |
THSD1
|
thrombospondin, type I, domain containing 1 |
| chr19_-_5680231 | 0.23 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr14_-_39417410 | 0.23 |
ENST00000557019.1
ENST00000556116.1 ENST00000554732.1 |
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr11_-_35287243 | 0.22 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_44588679 | 0.22 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr12_-_10251576 | 0.22 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr9_+_103189660 | 0.22 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr18_+_63417864 | 0.22 |
ENST00000536984.2
|
CDH7
|
cadherin 7, type 2 |
| chr9_+_112542591 | 0.21 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr1_-_71546690 | 0.21 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr2_-_148779106 | 0.21 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr4_-_40516560 | 0.20 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr10_-_111683183 | 0.20 |
ENST00000403138.2
ENST00000369683.1 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr7_-_105752971 | 0.20 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr4_-_76862117 | 0.20 |
ENST00000507956.1
ENST00000507187.2 ENST00000399497.3 ENST00000286733.4 |
NAAA
|
N-acylethanolamine acid amidase |
| chr10_-_111683308 | 0.20 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr9_+_127023704 | 0.20 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr21_-_34915147 | 0.20 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr15_-_74284558 | 0.19 |
ENST00000359750.4
ENST00000541638.1 ENST00000562453.1 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr6_+_88106840 | 0.19 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr2_-_44588694 | 0.19 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr12_+_28605426 | 0.18 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr2_-_44588624 | 0.18 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr21_+_39668831 | 0.17 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_-_186080012 | 0.17 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr16_-_8962200 | 0.16 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr14_+_56127989 | 0.16 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr10_-_105421427 | 0.16 |
ENST00000538130.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr21_+_45993606 | 0.15 |
ENST00000400374.3
|
KRTAP10-4
|
keratin associated protein 10-4 |
| chr17_-_7145475 | 0.15 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr17_-_79895154 | 0.14 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr1_+_144220127 | 0.14 |
ENST00000369373.5
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr1_+_10003486 | 0.14 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr14_+_56127960 | 0.13 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_32860020 | 0.13 |
ENST00000527163.1
ENST00000341071.7 ENST00000530485.1 ENST00000446293.2 ENST00000413080.1 ENST00000449308.1 ENST00000526031.1 ENST00000419121.2 ENST00000455895.2 |
BSDC1
|
BSD domain containing 1 |
| chr4_+_145567297 | 0.13 |
ENST00000434550.2
|
HHIP
|
hedgehog interacting protein |
| chr22_+_41763274 | 0.12 |
ENST00000406644.3
|
TEF
|
thyrotrophic embryonic factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of OSR1_OSR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.9 | 2.8 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.7 | 4.4 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.5 | 1.6 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.5 | 2.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.5 | 2.4 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.5 | 1.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.4 | 1.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.4 | 2.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.2 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 | 2.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.4 | 2.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 1.0 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.3 | 1.6 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.3 | 1.9 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.3 | 1.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.3 | 1.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.2 | 5.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.8 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 0.7 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 0.5 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 0.5 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.2 | 0.6 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.2 | 2.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 8.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 2.4 | GO:0042117 | monocyte activation(GO:0042117) cellular response to copper ion(GO:0071280) |
| 0.1 | 1.8 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 2.6 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 1.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 1.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 2.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.6 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.7 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.4 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 2.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 5.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 1.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 3.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.4 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 1.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 1.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 2.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.6 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.6 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 2.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 5.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 2.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 0.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 8.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 4.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 3.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 22.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.6 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 4.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.5 | 2.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.5 | 1.4 | GO:0016992 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.5 | 24.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.4 | 1.9 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 2.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 2.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 8.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 5.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.5 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.2 | 0.5 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 1.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.6 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 4.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.8 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 3.8 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 3.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 24.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |