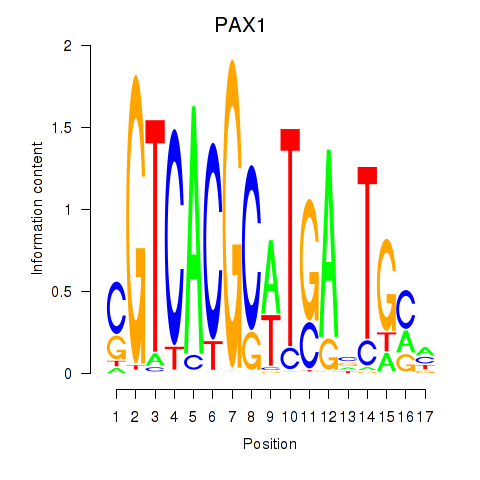
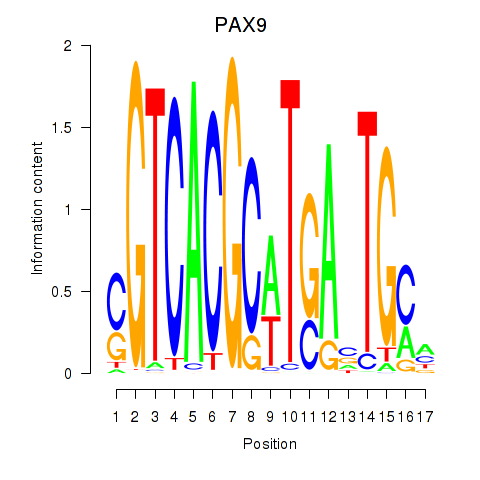
Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for PAX1_PAX9
Z-value: 1.02
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
PAX9
|
ENSG00000198807.8 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37131058_37131139 | 0.24 | 1.9e-01 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | -0.16 | 3.8e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_20566616 | 3.86 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr4_+_123747834 | 3.54 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr4_+_123747979 | 3.30 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr2_-_207630033 | 2.83 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr2_-_207629997 | 2.76 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr9_+_33750667 | 2.55 |
ENST00000457896.1
ENST00000342836.4 ENST00000429677.3 |
PRSS3
|
protease, serine, 3 |
| chr7_+_36429424 | 2.48 |
ENST00000396068.2
|
ANLN
|
anillin, actin binding protein |
| chr7_+_36429409 | 2.41 |
ENST00000265748.2
|
ANLN
|
anillin, actin binding protein |
| chr14_+_24540046 | 2.24 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr19_-_409134 | 2.17 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr9_+_33750515 | 2.12 |
ENST00000361005.5
|
PRSS3
|
protease, serine, 3 |
| chr6_-_137366163 | 2.11 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr5_+_55354822 | 1.78 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr9_-_104249400 | 1.65 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr18_-_47813940 | 1.44 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr19_-_18995029 | 1.42 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr11_+_61522844 | 1.31 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr18_-_47814032 | 1.30 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr4_-_171011084 | 1.29 |
ENST00000337664.4
|
AADAT
|
aminoadipate aminotransferase |
| chr9_+_33795533 | 1.27 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr18_+_39766626 | 1.25 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr9_-_124991124 | 1.22 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr3_+_195447738 | 1.11 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr7_-_22234381 | 1.09 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_171011323 | 1.08 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr2_-_175712270 | 1.06 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr17_+_39969183 | 1.01 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr2_-_175711978 | 0.99 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr8_-_23282820 | 0.98 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr8_-_23282797 | 0.95 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr19_+_14184370 | 0.88 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr1_-_162838551 | 0.84 |
ENST00000367910.1
ENST00000367912.2 ENST00000367911.2 |
C1orf110
|
chromosome 1 open reading frame 110 |
| chr4_-_163085141 | 0.81 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr8_-_23261589 | 0.78 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr20_-_17641097 | 0.78 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1 |
| chr11_+_63870660 | 0.72 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr19_-_5838734 | 0.69 |
ENST00000532464.1
ENST00000528505.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr19_-_5838768 | 0.68 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr17_+_39968926 | 0.68 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr17_-_3461092 | 0.67 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr10_-_99393242 | 0.65 |
ENST00000370635.3
ENST00000478953.1 ENST00000335628.3 |
MORN4
|
MORN repeat containing 4 |
| chr10_-_99393208 | 0.54 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr6_+_159291090 | 0.50 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr12_+_57828521 | 0.50 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr7_-_19157248 | 0.47 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr19_+_48337701 | 0.47 |
ENST00000535362.1
|
TPRX2P
|
tetra-peptide repeat homeobox 2 pseudogene |
| chrX_-_132091284 | 0.46 |
ENST00000370833.2
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr3_-_57113281 | 0.46 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr11_+_6947720 | 0.45 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr17_+_15848231 | 0.44 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr1_-_204165610 | 0.38 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr11_-_67397371 | 0.34 |
ENST00000376693.2
ENST00000301490.4 |
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr9_+_96793076 | 0.33 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_153707545 | 0.32 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr6_+_3118926 | 0.29 |
ENST00000380379.5
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase) |
| chr2_-_39414848 | 0.29 |
ENST00000451199.1
|
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr9_-_21142144 | 0.27 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr12_+_58138800 | 0.24 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr22_+_39745930 | 0.22 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr9_+_6716478 | 0.21 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr12_-_122658746 | 0.20 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr14_+_64854958 | 0.19 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr6_+_34857019 | 0.19 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr17_-_39968406 | 0.17 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr6_+_159290917 | 0.14 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr2_+_234160340 | 0.14 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr14_+_39703112 | 0.13 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr6_+_132891461 | 0.11 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr4_+_75230853 | 0.09 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr16_-_84076241 | 0.08 |
ENST00000568178.1
|
SLC38A8
|
solute carrier family 38, member 8 |
| chr2_+_234160217 | 0.08 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr5_+_133842243 | 0.07 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr13_+_21141270 | 0.06 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr13_+_98086445 | 0.05 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr12_-_101801505 | 0.01 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr1_+_213031570 | 0.00 |
ENST00000366971.4
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 1.6 | 4.9 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.7 | 2.7 | GO:0018277 | protein deamination(GO:0018277) |
| 0.5 | 1.4 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.4 | 1.7 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.4 | 2.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.3 | 5.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 5.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.5 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 1.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.4 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.7 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 2.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 2.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 2.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 2.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 3.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 5.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.7 | 2.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.6 | 5.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 2.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 2.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 3.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 1.4 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.2 | 7.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 2.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 7.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 3.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |