Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for PAX6
Z-value: 1.39
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg19_v2_chr11_-_31839488_31839515 | 0.75 | 8.8e-07 | Click! |
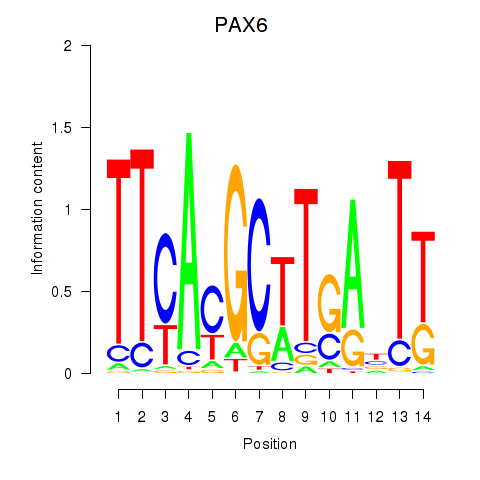
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35820064 | 2.84 |
ENST00000341773.6
ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22
|
CD22 molecule |
| chr8_+_21882244 | 2.17 |
ENST00000289820.6
ENST00000381530.5 |
NPM2
|
nucleophosmin/nucleoplasmin 2 |
| chr19_-_22806764 | 2.06 |
ENST00000598042.1
|
AC011516.2
|
AC011516.2 |
| chr12_-_45270077 | 1.97 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr4_+_115519577 | 1.90 |
ENST00000310836.6
|
UGT8
|
UDP glycosyltransferase 8 |
| chr12_-_45270151 | 1.86 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr15_+_51973550 | 1.77 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr15_+_51973680 | 1.75 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr20_-_44144249 | 1.64 |
ENST00000217428.6
|
SPINT3
|
serine peptidase inhibitor, Kunitz type, 3 |
| chr12_-_45269430 | 1.62 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr19_+_50936142 | 1.56 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr15_+_51973708 | 1.48 |
ENST00000558709.1
|
SCG3
|
secretogranin III |
| chr4_+_76995855 | 1.46 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr14_-_36645674 | 1.43 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr12_-_70093111 | 1.43 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr10_-_118765081 | 1.42 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr2_-_158182410 | 1.37 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_-_158182322 | 1.37 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr18_+_18822185 | 1.34 |
ENST00000424526.1
ENST00000400483.4 ENST00000431264.1 |
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr10_-_118764862 | 1.33 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr12_-_45269769 | 1.33 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chrX_-_92928557 | 1.29 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr9_+_87283430 | 1.23 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_+_2005425 | 1.22 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chrX_-_130423200 | 1.22 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr12_-_122107549 | 1.19 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr11_-_18813353 | 1.18 |
ENST00000358540.2
ENST00000396171.4 ENST00000396167.2 |
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr12_+_59194154 | 1.18 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chrX_-_130423386 | 1.17 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr1_-_99470558 | 1.17 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr16_+_77822427 | 1.13 |
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like |
| chr13_+_96085847 | 1.12 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr17_+_58499844 | 1.11 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chrX_-_130423240 | 1.09 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr1_-_184006829 | 1.08 |
ENST00000361927.4
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr5_+_322685 | 1.06 |
ENST00000510400.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr12_-_70093065 | 1.06 |
ENST00000553096.1
|
BEST3
|
bestrophin 3 |
| chr11_-_18813110 | 1.06 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr12_-_70093190 | 1.05 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
| chr2_+_196521458 | 1.03 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr16_-_51185172 | 1.01 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr12_-_45269251 | 1.01 |
ENST00000553120.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr2_-_154335300 | 0.99 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr5_+_17404114 | 0.99 |
ENST00000508677.1
|
RP11-321E2.3
|
RP11-321E2.3 |
| chr9_-_140142222 | 0.99 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr4_-_174320687 | 0.99 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chrX_+_10126488 | 0.97 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr4_+_70916119 | 0.97 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr2_+_196521903 | 0.97 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr8_-_95487272 | 0.96 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr18_+_13611431 | 0.95 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr8_-_18711866 | 0.94 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr12_+_58937907 | 0.90 |
ENST00000546977.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr22_-_29457832 | 0.90 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr9_-_116861337 | 0.89 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr2_+_196521845 | 0.88 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr11_+_33563821 | 0.88 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr12_-_70093235 | 0.87 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr11_-_117688216 | 0.86 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr5_+_17404147 | 0.85 |
ENST00000507730.1
|
RP11-321E2.3
|
RP11-321E2.3 |
| chr2_-_158182105 | 0.85 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chrX_-_124097620 | 0.84 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr10_+_18240753 | 0.84 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr10_+_18240834 | 0.83 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr16_-_51185149 | 0.81 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr17_-_8661860 | 0.81 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr6_+_30844192 | 0.80 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_140981456 | 0.80 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr19_-_57347415 | 0.79 |
ENST00000601070.1
|
ZIM2
|
zinc finger, imprinted 2 |
| chr11_-_132812987 | 0.79 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr10_+_18240814 | 0.79 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr6_+_26156551 | 0.77 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr12_+_111284764 | 0.77 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr5_+_321810 | 0.76 |
ENST00000514523.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr2_+_168675182 | 0.75 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_-_73736511 | 0.75 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr3_+_167582561 | 0.73 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr8_-_79470728 | 0.72 |
ENST00000522807.1
ENST00000519242.1 ENST00000522302.1 |
RP11-594N15.2
|
RP11-594N15.2 |
| chrX_-_80457385 | 0.72 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr12_-_85306594 | 0.72 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr4_-_65275162 | 0.71 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr6_-_26043885 | 0.70 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr22_+_39745930 | 0.70 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr15_+_52043758 | 0.70 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr4_+_114066764 | 0.69 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr9_-_140142181 | 0.69 |
ENST00000484720.1
|
FAM166A
|
family with sequence similarity 166, member A |
| chr4_-_163085141 | 0.68 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr17_-_32906388 | 0.67 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr8_-_19615435 | 0.67 |
ENST00000523262.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr17_-_58499766 | 0.67 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr11_+_33563618 | 0.67 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr5_-_135231516 | 0.66 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
| chr15_-_56757329 | 0.66 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr1_-_36906474 | 0.66 |
ENST00000433045.2
|
OSCP1
|
organic solute carrier partner 1 |
| chr12_-_78753496 | 0.66 |
ENST00000548512.1
|
RP11-38F22.1
|
RP11-38F22.1 |
| chr18_+_18822216 | 0.66 |
ENST00000269218.6
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr3_+_51989330 | 0.65 |
ENST00000322241.4
|
GPR62
|
G protein-coupled receptor 62 |
| chr10_-_16563870 | 0.65 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr3_-_36781352 | 0.65 |
ENST00000416516.2
|
DCLK3
|
doublecortin-like kinase 3 |
| chr5_-_68339648 | 0.65 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr12_-_85306562 | 0.64 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_-_104597286 | 0.64 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr1_+_174670143 | 0.63 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_+_104068562 | 0.63 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr12_-_70093162 | 0.62 |
ENST00000551160.1
|
BEST3
|
bestrophin 3 |
| chr8_+_30496078 | 0.61 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr1_-_185597619 | 0.61 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr4_-_87281224 | 0.59 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_142286887 | 0.59 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr4_-_163085107 | 0.59 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr5_+_57787254 | 0.59 |
ENST00000502276.1
ENST00000396776.2 ENST00000511930.1 |
GAPT
|
GRB2-binding adaptor protein, transmembrane |
| chr19_-_54327542 | 0.58 |
ENST00000391775.3
ENST00000324134.6 ENST00000535162.1 ENST00000351894.4 ENST00000354278.3 ENST00000391773.1 ENST00000345770.5 ENST00000391772.1 |
NLRP12
|
NLR family, pyrin domain containing 12 |
| chr12_+_111284805 | 0.58 |
ENST00000552694.1
|
CCDC63
|
coiled-coil domain containing 63 |
| chr7_-_150754935 | 0.58 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr5_-_160279207 | 0.57 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr8_+_21916710 | 0.57 |
ENST00000523266.1
ENST00000519907.1 |
DMTN
|
dematin actin binding protein |
| chr15_+_68924327 | 0.57 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr8_-_19615382 | 0.57 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr20_+_32250079 | 0.57 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr12_+_83080724 | 0.56 |
ENST00000548305.1
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_+_73771844 | 0.56 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr9_+_129097520 | 0.56 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr5_+_140254884 | 0.56 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr10_+_90354503 | 0.56 |
ENST00000531458.1
|
LIPJ
|
lipase, family member J |
| chr2_+_191334212 | 0.55 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr14_-_47812321 | 0.55 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr1_+_218683438 | 0.55 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr5_+_67485704 | 0.55 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr9_+_93589734 | 0.55 |
ENST00000375746.1
|
SYK
|
spleen tyrosine kinase |
| chr4_-_798689 | 0.55 |
ENST00000504062.1
|
CPLX1
|
complexin 1 |
| chr21_-_26797019 | 0.54 |
ENST00000440205.1
|
LINC00158
|
long intergenic non-protein coding RNA 158 |
| chr16_-_71264558 | 0.53 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr8_+_21916680 | 0.53 |
ENST00000358242.3
ENST00000415253.1 |
DMTN
|
dematin actin binding protein |
| chr19_-_14945933 | 0.53 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr1_+_84609944 | 0.53 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_+_123884038 | 0.53 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr10_+_35894338 | 0.52 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chr1_+_174669653 | 0.52 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_13835147 | 0.52 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr9_+_129097479 | 0.51 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr5_+_322733 | 0.51 |
ENST00000515206.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr5_+_93954039 | 0.51 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr17_+_57297807 | 0.50 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr8_+_121457642 | 0.50 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr12_-_2045694 | 0.48 |
ENST00000418006.1
|
LINC00940
|
long intergenic non-protein coding RNA 940 |
| chr7_-_80551671 | 0.48 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr18_-_37380230 | 0.48 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr8_-_27469383 | 0.48 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr8_-_11853819 | 0.48 |
ENST00000526438.1
ENST00000382205.4 |
DEFB134
|
defensin, beta 134 |
| chr6_+_147527103 | 0.47 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr4_-_65275100 | 0.47 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr12_-_3982548 | 0.47 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr3_-_196910477 | 0.46 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_-_175869936 | 0.46 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr14_+_66578299 | 0.46 |
ENST00000554187.1
ENST00000556662.1 ENST00000556291.1 ENST00000557723.1 ENST00000557050.1 |
RP11-783L4.1
|
RP11-783L4.1 |
| chr1_+_163291732 | 0.46 |
ENST00000271452.3
|
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr2_+_196522032 | 0.45 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_-_106449656 | 0.45 |
ENST00000372466.4
ENST00000421752.1 ENST00000372461.3 |
NUP62CL
|
nucleoporin 62kDa C-terminal like |
| chr7_-_30726065 | 0.44 |
ENST00000341843.4
|
CRHR2
|
corticotropin releasing hormone receptor 2 |
| chr10_+_91092241 | 0.44 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr14_+_22320634 | 0.43 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr5_-_159766528 | 0.43 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr5_-_156593273 | 0.43 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71, member B |
| chr2_+_132160448 | 0.43 |
ENST00000437751.1
|
AC073869.19
|
long intergenic non-protein coding RNA 1120 |
| chr4_+_40751914 | 0.43 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr13_+_24844979 | 0.43 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr10_+_116524547 | 0.42 |
ENST00000436932.1
|
RP11-106M7.1
|
RP11-106M7.1 |
| chr18_+_32173276 | 0.42 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr2_-_85645545 | 0.42 |
ENST00000409275.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr14_+_21569245 | 0.42 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr18_+_13611763 | 0.41 |
ENST00000585931.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr4_+_129349188 | 0.41 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr1_-_89458287 | 0.40 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr10_-_17243579 | 0.40 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr19_-_10047219 | 0.40 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr19_-_14911023 | 0.40 |
ENST00000248073.2
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1 |
| chr12_+_78224667 | 0.39 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr11_+_26353664 | 0.39 |
ENST00000531646.1
|
ANO3
|
anoctamin 3 |
| chr2_-_175870085 | 0.38 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr7_-_152373216 | 0.38 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr7_-_97501706 | 0.38 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr2_+_29320571 | 0.38 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr2_-_86116020 | 0.38 |
ENST00000525834.2
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr9_+_129278696 | 0.38 |
ENST00000454034.1
|
RP11-205K6.1
|
RP11-205K6.1 |
| chr19_-_51920873 | 0.38 |
ENST00000441969.3
ENST00000525998.1 ENST00000436984.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr19_+_49891475 | 0.37 |
ENST00000447857.3
|
CCDC155
|
coiled-coil domain containing 155 |
| chrX_+_144908928 | 0.37 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr12_+_83080659 | 0.37 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr6_-_2634820 | 0.37 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr1_+_15668240 | 0.37 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr1_-_156269428 | 0.37 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr22_-_30968813 | 0.37 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_+_42037433 | 0.36 |
ENST00000599316.1
ENST00000599770.1 |
AC006129.1
|
AC006129.1 |
| chr7_-_97501733 | 0.36 |
ENST00000444334.1
ENST00000422745.1 ENST00000394308.3 ENST00000451771.1 ENST00000175506.4 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr19_-_51920835 | 0.36 |
ENST00000442846.3
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr18_-_45663666 | 0.36 |
ENST00000535628.2
|
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr4_+_139694701 | 0.36 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr2_-_60580001 | 0.35 |
ENST00000457668.1
|
AC007381.3
|
AC007381.3 |
| chr12_-_2986107 | 0.35 |
ENST00000359843.3
ENST00000342628.2 ENST00000361953.3 |
FOXM1
|
forkhead box M1 |
| chr1_+_172745006 | 0.35 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.4 | 7.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 1.8 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.3 | 3.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.7 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 1.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.5 | GO:0045401 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 2.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.4 | GO:0015820 | leucine transport(GO:0015820) |
| 0.1 | 0.7 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.6 | GO:0032661 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 1.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.3 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.1 | 1.2 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.8 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 1.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.5 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.6 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.5 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.4 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.2 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 1.2 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 6.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0051446 | positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.6 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.7 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.3 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 4.5 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.4 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.5 | 2.3 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.3 | 3.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 1.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 1.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.8 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 5.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.0 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 3.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 3.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 2.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.4 | 3.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 0.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 1.7 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.6 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 1.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.8 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 5.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.4 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 1.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 1.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 6.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 7.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 5.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |