Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for PGR
Z-value: 1.79
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_100999775_100999801 | 0.54 | 1.5e-03 | Click! |
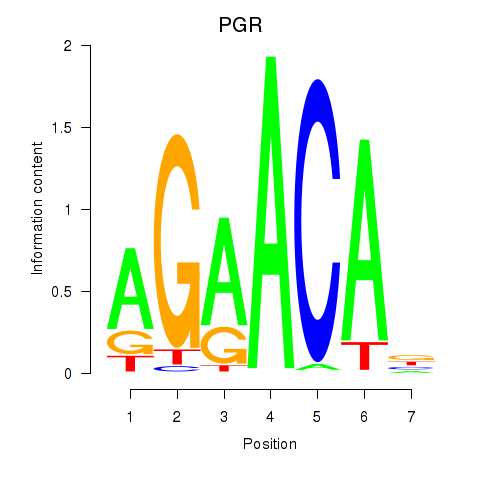
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_54305101 | 4.23 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr4_-_88450612 | 3.94 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr19_+_30863271 | 3.76 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr4_-_88450535 | 3.74 |
ENST00000541496.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr4_-_88450372 | 3.48 |
ENST00000543631.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr4_-_88450595 | 3.44 |
ENST00000434434.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr20_-_52612705 | 3.37 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_-_88450511 | 3.34 |
ENST00000458304.2
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr4_-_154710210 | 3.32 |
ENST00000274063.4
|
SFRP2
|
secreted frizzled-related protein 2 |
| chr15_-_49255632 | 3.27 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr8_-_22089845 | 3.14 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr20_-_52612468 | 3.02 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chrX_-_132095419 | 2.99 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr12_-_88974236 | 2.78 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr10_-_49482907 | 2.61 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr8_-_26371608 | 2.55 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr11_+_27062860 | 2.50 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_87028478 | 2.44 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr20_+_58203664 | 2.39 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr5_-_160279207 | 2.36 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr3_-_139195350 | 2.25 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr8_+_35649365 | 2.20 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr2_+_73144604 | 2.18 |
ENST00000258106.6
|
EMX1
|
empty spiracles homeobox 1 |
| chrX_+_38420623 | 2.11 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chrX_-_21776281 | 2.08 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr17_-_42988004 | 2.07 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr3_-_58563094 | 2.05 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr19_-_42947121 | 2.05 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr12_-_71551652 | 2.03 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_197237352 | 2.03 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr18_+_31185530 | 1.98 |
ENST00000586327.1
|
ASXL3
|
additional sex combs like 3 (Drosophila) |
| chr1_-_26394114 | 1.95 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr15_-_29745003 | 1.94 |
ENST00000560082.1
|
FAM189A1
|
family with sequence similarity 189, member A1 |
| chr18_+_580367 | 1.93 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr2_-_183387064 | 1.88 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr15_+_48483736 | 1.88 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_+_16370271 | 1.83 |
ENST00000375679.4
|
CLCNKB
|
chloride channel, voltage-sensitive Kb |
| chr15_-_65360276 | 1.81 |
ENST00000421977.3
|
RASL12
|
RAS-like, family 12 |
| chr2_-_175711978 | 1.79 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr9_+_87285539 | 1.79 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_+_127615733 | 1.77 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr18_-_44336998 | 1.76 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr13_+_102104980 | 1.75 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr11_-_75379612 | 1.74 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr18_+_32173276 | 1.71 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr13_+_102104952 | 1.69 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr16_-_66952779 | 1.69 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_+_69865866 | 1.67 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chrX_+_70364667 | 1.67 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr1_-_32210275 | 1.66 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr14_+_29299437 | 1.65 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr17_+_11144580 | 1.62 |
ENST00000441885.3
ENST00000432116.3 ENST00000409168.3 |
SHISA6
|
shisa family member 6 |
| chr7_+_123241908 | 1.61 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr5_+_140261703 | 1.60 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr20_+_33589048 | 1.59 |
ENST00000446156.1
ENST00000453028.1 ENST00000435272.1 ENST00000433934.2 ENST00000456649.1 |
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr19_-_36247910 | 1.59 |
ENST00000587965.1
ENST00000004982.3 |
HSPB6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr10_+_68685764 | 1.58 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_125985620 | 1.56 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr9_+_1051481 | 1.56 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr17_+_37824411 | 1.55 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr2_-_175711924 | 1.54 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr3_+_158991025 | 1.53 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_+_58497274 | 1.52 |
ENST00000564207.1
|
NDRG4
|
NDRG family member 4 |
| chrX_-_114253536 | 1.52 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr16_-_66952742 | 1.52 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_+_72926145 | 1.51 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr15_-_76005170 | 1.50 |
ENST00000308508.5
|
CSPG4
|
chondroitin sulfate proteoglycan 4 |
| chr4_-_87770416 | 1.50 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr5_-_121413974 | 1.50 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr4_+_79567233 | 1.49 |
ENST00000514130.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr11_-_75379479 | 1.48 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr8_-_11325047 | 1.48 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr6_-_47010061 | 1.46 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr2_-_183387283 | 1.46 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183387430 | 1.45 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_+_178482212 | 1.45 |
ENST00000319416.2
ENST00000258298.2 ENST00000367643.3 ENST00000367642.3 |
TEX35
|
testis expressed 35 |
| chr15_-_58306295 | 1.44 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr1_-_166944652 | 1.44 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr8_+_70476088 | 1.44 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr2_-_158182410 | 1.43 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr3_-_149051194 | 1.43 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr11_-_126810521 | 1.42 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr4_-_139163491 | 1.42 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_-_205391178 | 1.37 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr3_+_173113898 | 1.37 |
ENST00000423427.1
|
NLGN1
|
neuroligin 1 |
| chr18_-_3880051 | 1.37 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_158182322 | 1.36 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_-_134326009 | 1.36 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr2_+_48796120 | 1.36 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr1_-_166944561 | 1.35 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr9_-_73736511 | 1.35 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_+_95286151 | 1.35 |
ENST00000467909.1
ENST00000422520.2 ENST00000532427.1 |
SLC44A3
|
solute carrier family 44, member 3 |
| chr13_-_45768841 | 1.34 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr17_+_37824217 | 1.33 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chrX_+_134887233 | 1.33 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr11_-_88796803 | 1.32 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chrX_+_135570046 | 1.31 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr2_+_232135245 | 1.31 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr5_+_140797296 | 1.30 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr10_-_118429461 | 1.30 |
ENST00000588184.1
ENST00000369210.3 |
C10orf82
|
chromosome 10 open reading frame 82 |
| chr6_+_30852738 | 1.29 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_79258547 | 1.29 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr16_+_86600857 | 1.28 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr4_-_90759440 | 1.28 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_-_19144243 | 1.28 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr15_-_88799661 | 1.27 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr6_+_30853002 | 1.26 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_-_23876801 | 1.26 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr1_-_234667504 | 1.26 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr17_+_65027509 | 1.25 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr9_-_34665983 | 1.25 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr7_+_30960915 | 1.25 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_-_175711133 | 1.25 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr11_-_26744908 | 1.24 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_+_58003935 | 1.24 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr2_-_180427304 | 1.22 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr6_+_74405501 | 1.22 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr15_-_88799384 | 1.22 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr2_-_183106641 | 1.21 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_180650271 | 1.21 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr8_-_52721975 | 1.21 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr3_-_179754806 | 1.21 |
ENST00000485199.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr17_-_15168624 | 1.21 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr1_+_162335197 | 1.21 |
ENST00000431696.1
|
RP11-565P22.6
|
Uncharacterized protein |
| chr1_-_145039835 | 1.20 |
ENST00000533259.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_-_46922659 | 1.20 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr10_+_97709725 | 1.20 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr3_-_179754556 | 1.18 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr13_-_103053946 | 1.17 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr19_+_46806856 | 1.16 |
ENST00000300862.3
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr1_+_172422026 | 1.16 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr20_-_44168046 | 1.14 |
ENST00000372665.3
ENST00000372670.3 ENST00000600168.1 |
WFDC6
|
WAP four-disulfide core domain 6 |
| chr7_-_92855762 | 1.13 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_59477233 | 1.13 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr3_-_48057890 | 1.13 |
ENST00000434267.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_108487245 | 1.13 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_+_173686303 | 1.13 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr3_-_149051444 | 1.12 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr1_-_201391149 | 1.12 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr4_+_128802016 | 1.11 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr12_-_81763184 | 1.09 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr18_+_43307294 | 1.07 |
ENST00000590246.1
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr1_-_112903150 | 1.07 |
ENST00000427290.1
|
RP5-965F6.2
|
RP5-965F6.2 |
| chr7_+_99202003 | 1.06 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr1_-_156828810 | 1.06 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr6_+_74405804 | 1.05 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr5_+_102201430 | 1.04 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_179754733 | 1.03 |
ENST00000472994.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr16_-_3350614 | 1.03 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr17_+_7308172 | 1.02 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr9_-_140196703 | 1.01 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr5_+_137419581 | 1.01 |
ENST00000506684.1
ENST00000504809.1 ENST00000398754.1 |
WNT8A
|
wingless-type MMTV integration site family, member 8A |
| chr19_+_51153045 | 1.00 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr7_-_123673471 | 0.99 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr17_+_67590125 | 0.98 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr7_+_105603738 | 0.98 |
ENST00000541203.1
|
CDHR3
|
cadherin-related family member 3 |
| chr3_+_147795932 | 0.97 |
ENST00000490465.1
|
RP11-639B1.1
|
RP11-639B1.1 |
| chr5_-_179107975 | 0.97 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr3_-_114343039 | 0.97 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_40549984 | 0.96 |
ENST00000457989.1
|
AC079630.2
|
AC079630.2 |
| chr7_+_73442487 | 0.96 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr16_-_54882145 | 0.96 |
ENST00000561591.1
|
RP11-1136G4.1
|
RP11-1136G4.1 |
| chr2_-_167350747 | 0.96 |
ENST00000419992.1
|
SCN7A
|
sodium channel, voltage-gated, type VII, alpha subunit |
| chr7_+_105603683 | 0.95 |
ENST00000317716.9
|
CDHR3
|
cadherin-related family member 3 |
| chr3_-_179754706 | 0.95 |
ENST00000465751.1
ENST00000467460.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr1_-_145039771 | 0.94 |
ENST00000493130.2
ENST00000532801.1 ENST00000478649.2 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr14_-_21056121 | 0.94 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr5_-_38557561 | 0.94 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr7_+_79763271 | 0.93 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr19_+_42254885 | 0.93 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr1_-_61719160 | 0.93 |
ENST00000442027.1
ENST00000596354.1 ENST00000596404.1 |
RP5-833A20.1
|
RP5-833A20.1 |
| chr11_+_125034640 | 0.92 |
ENST00000542175.1
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr16_+_202686 | 0.92 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr1_+_111770278 | 0.91 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr12_-_11422630 | 0.91 |
ENST00000381842.3
ENST00000538488.1 |
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr8_+_142524738 | 0.91 |
ENST00000427937.1
|
AC138647.1
|
Uncharacterized protein |
| chr8_-_87755878 | 0.91 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr10_-_97200772 | 0.91 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr7_-_14026123 | 0.90 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr1_+_178482262 | 0.90 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr6_-_76203454 | 0.90 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_-_110933663 | 0.89 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr2_+_111562758 | 0.89 |
ENST00000433706.1
|
ACOXL
|
acyl-CoA oxidase-like |
| chr5_+_72469014 | 0.89 |
ENST00000296776.5
|
TMEM174
|
transmembrane protein 174 |
| chr1_-_145039949 | 0.89 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_111770232 | 0.88 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chrX_+_101478829 | 0.88 |
ENST00000372763.1
ENST00000372758.1 |
NXF2
|
nuclear RNA export factor 2 |
| chr11_-_62689046 | 0.88 |
ENST00000306960.3
ENST00000543973.1 |
CHRM1
|
cholinergic receptor, muscarinic 1 |
| chr3_+_25469802 | 0.88 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chrX_+_109602039 | 0.87 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr2_-_216300784 | 0.87 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr6_-_150212029 | 0.87 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr17_-_46691990 | 0.86 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr18_+_43304092 | 0.86 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr2_-_158182105 | 0.86 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr8_+_65285851 | 0.85 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr18_+_6729725 | 0.84 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr7_+_105603699 | 0.84 |
ENST00000478080.1
|
CDHR3
|
cadherin-related family member 3 |
| chr4_+_100432161 | 0.83 |
ENST00000326581.4
ENST00000514652.1 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr1_-_85870177 | 0.82 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.9 | 2.8 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.7 | 2.9 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.6 | 2.5 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.5 | 1.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.5 | 3.0 | GO:0018032 | protein amidation(GO:0018032) |
| 0.5 | 3.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.3 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.4 | 4.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 1.2 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.4 | 4.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 2.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 1.1 | GO:1901207 | mammary placode formation(GO:0060596) regulation of heart looping(GO:1901207) |
| 0.4 | 4.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 1.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 1.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 1.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.3 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 6.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.8 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.3 | 2.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 0.9 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.3 | 2.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 1.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 1.1 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 2.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 1.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.3 | 1.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.3 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.3 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 | 1.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 3.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 1.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 3.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 2.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 2.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 2.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 4.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 2.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.5 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 0.4 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 3.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 2.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.9 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 1.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 4.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 2.0 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 1.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 1.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.5 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 2.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.3 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.8 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 1.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 1.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.6 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 1.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 1.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.6 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 8.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.5 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 1.5 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 1.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 17.1 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.9 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.6 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.4 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.0 | 0.0 | GO:0061551 | trigeminal ganglion development(GO:0061551) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 1.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 2.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 0.8 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.3 | 3.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 1.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 1.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.0 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0097386 | voltage-gated sodium channel complex(GO:0001518) glial cell projection(GO:0097386) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 4.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 15.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 7.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 3.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.9 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 6.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.9 | 6.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.7 | 3.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.7 | 2.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 1.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.6 | 2.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 3.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 4.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 1.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 3.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 3.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 1.8 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 1.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.3 | 2.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 1.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 1.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 3.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.2 | 2.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 4.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 14.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.9 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 4.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 2.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 3.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 2.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 2.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.0 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.0 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 19.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |