Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for PKNOX2
Z-value: 0.56
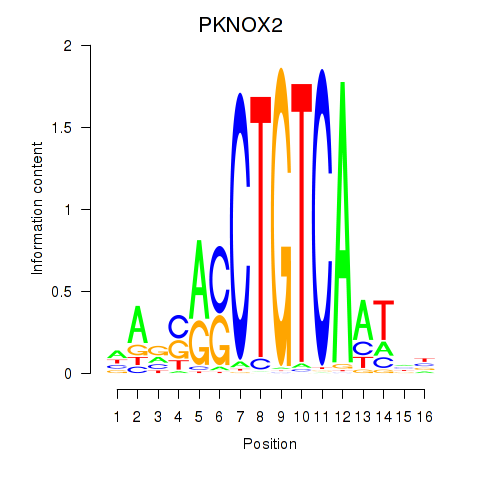
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX2 | hg19_v2_chr11_+_125034605_125034636 | 0.14 | 4.6e-01 | Click! |
Activity profile of PKNOX2 motif
Sorted Z-values of PKNOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_31674639 | 0.85 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr21_-_34185989 | 0.68 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr1_-_201081579 | 0.63 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr21_-_34185944 | 0.59 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_-_211179883 | 0.59 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr14_+_22958441 | 0.58 |
ENST00000390488.1
|
TRAJ49
|
T cell receptor alpha joining 49 |
| chr5_-_58882219 | 0.57 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_-_42452063 | 0.57 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr2_+_113321939 | 0.53 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr21_-_34186006 | 0.52 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr17_-_29151686 | 0.48 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr17_-_29151794 | 0.48 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chrX_-_153095813 | 0.45 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr21_-_32931290 | 0.44 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr14_-_21994337 | 0.43 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_66258846 | 0.43 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr18_+_54318566 | 0.42 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr11_-_71810258 | 0.42 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr6_-_114664180 | 0.41 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr8_+_11666649 | 0.41 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr14_-_21994525 | 0.40 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr9_-_132805430 | 0.39 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chrX_-_39923656 | 0.38 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr6_-_84418841 | 0.38 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr9_-_79307096 | 0.37 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr18_+_54318616 | 0.36 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr7_-_100253993 | 0.36 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr6_-_84418860 | 0.36 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr5_-_139283982 | 0.33 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr5_-_132073111 | 0.33 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr22_-_31885514 | 0.33 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr1_-_247171347 | 0.32 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chrX_-_153775426 | 0.31 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr18_+_54318893 | 0.30 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chrX_-_77225135 | 0.29 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr22_+_20905269 | 0.29 |
ENST00000457322.1
|
MED15
|
mediator complex subunit 15 |
| chr1_-_54872059 | 0.29 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr1_+_161736072 | 0.28 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_+_145549203 | 0.28 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr6_+_24126350 | 0.28 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr2_-_74780176 | 0.27 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr18_+_3449330 | 0.27 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_101395936 | 0.27 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr4_-_46391805 | 0.27 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr6_+_26124373 | 0.26 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr17_+_8316442 | 0.26 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr12_-_62653903 | 0.26 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr2_-_219524193 | 0.26 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
| chr3_+_140981456 | 0.25 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr11_+_12132117 | 0.25 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr15_+_34260921 | 0.25 |
ENST00000560035.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr6_-_101329191 | 0.25 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr17_+_38474489 | 0.25 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr1_-_179112173 | 0.24 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_-_72358001 | 0.24 |
ENST00000375366.3
|
BTBD17
|
BTB (POZ) domain containing 17 |
| chr2_-_242842587 | 0.24 |
ENST00000404031.1
|
AC131097.4
|
Protein LOC285095 |
| chr22_+_20905286 | 0.23 |
ENST00000428629.1
|
MED15
|
mediator complex subunit 15 |
| chr7_-_156803329 | 0.23 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr9_-_123639600 | 0.23 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr1_-_179112189 | 0.23 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr15_-_61521495 | 0.22 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr11_-_46867780 | 0.22 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr6_-_32140886 | 0.22 |
ENST00000395496.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr17_+_78518617 | 0.22 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr11_-_132812987 | 0.22 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr6_-_84418738 | 0.22 |
ENST00000519779.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_46391567 | 0.21 |
ENST00000507460.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr22_+_20905422 | 0.21 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr3_+_46742823 | 0.21 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr1_+_181057638 | 0.21 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr17_-_4046257 | 0.21 |
ENST00000381638.2
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1 |
| chr6_+_37225540 | 0.20 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr2_-_74779744 | 0.20 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr7_-_104909435 | 0.20 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr6_-_84418432 | 0.20 |
ENST00000519825.1
ENST00000523484.2 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr19_+_57050317 | 0.20 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr3_-_52001448 | 0.20 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr19_-_6424783 | 0.20 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr1_-_35658736 | 0.20 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr8_+_31496809 | 0.20 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr12_-_121734489 | 0.19 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr11_-_132813566 | 0.19 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr17_-_26220366 | 0.19 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr6_-_101329157 | 0.19 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr19_+_36142147 | 0.19 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr22_+_17082732 | 0.18 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr16_-_1275257 | 0.18 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr10_+_94608245 | 0.17 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr13_+_33160553 | 0.17 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr2_-_61765732 | 0.16 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr2_+_113033164 | 0.16 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr18_-_67872891 | 0.15 |
ENST00000454359.1
ENST00000437017.1 |
RTTN
|
rotatin |
| chr16_-_3030407 | 0.15 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr5_-_127873896 | 0.15 |
ENST00000502468.1
|
FBN2
|
fibrillin 2 |
| chr17_-_35969409 | 0.14 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chrX_+_41548220 | 0.14 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr4_-_149365827 | 0.14 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr6_+_110299501 | 0.14 |
ENST00000414000.2
|
GPR6
|
G protein-coupled receptor 6 |
| chr12_+_69201923 | 0.14 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr15_+_96904487 | 0.14 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr16_+_50776021 | 0.14 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr10_+_28822636 | 0.14 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr15_+_59908633 | 0.13 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_+_113263199 | 0.13 |
ENST00000361886.3
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
| chr9_-_123639304 | 0.13 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr20_+_43514320 | 0.13 |
ENST00000372839.3
ENST00000428262.1 ENST00000445830.1 |
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chr4_-_46391367 | 0.13 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr1_+_22778337 | 0.13 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr22_+_30279144 | 0.13 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr11_-_132813627 | 0.13 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr4_-_157892055 | 0.13 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr3_-_52719810 | 0.12 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr6_+_108881012 | 0.12 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr22_-_30234218 | 0.12 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_-_1782625 | 0.12 |
ENST00000438213.1
|
CTSD
|
cathepsin D |
| chr9_-_99637820 | 0.12 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr3_-_3221358 | 0.12 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr18_+_30349623 | 0.12 |
ENST00000426194.1
|
AC012123.1
|
Uncharacterized protein |
| chr2_-_61765315 | 0.11 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr4_+_3076388 | 0.11 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr17_+_39868577 | 0.11 |
ENST00000329402.3
|
GAST
|
gastrin |
| chr6_+_29910301 | 0.11 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr6_+_138188551 | 0.11 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr16_-_30457048 | 0.11 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr19_+_10828724 | 0.11 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr5_-_81046922 | 0.11 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr2_-_190446738 | 0.11 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr10_+_70661014 | 0.11 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chrX_+_41548259 | 0.11 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr17_-_27405875 | 0.11 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr6_+_150070831 | 0.10 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr6_+_42123141 | 0.10 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr6_-_149867122 | 0.10 |
ENST00000253329.2
|
PPIL4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr5_-_81046841 | 0.10 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr1_-_100598444 | 0.09 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr6_+_150070857 | 0.09 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr11_-_74660159 | 0.08 |
ENST00000527087.1
ENST00000321448.8 ENST00000340360.6 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr14_-_69619689 | 0.08 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr19_+_10828795 | 0.08 |
ENST00000389253.4
ENST00000355667.6 ENST00000408974.4 |
DNM2
|
dynamin 2 |
| chrX_+_2924654 | 0.08 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr2_+_64681219 | 0.08 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr9_-_73477826 | 0.08 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_-_111741994 | 0.07 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr16_+_19297047 | 0.07 |
ENST00000493231.1
|
CLEC19A
|
C-type lectin domain family 19, member A |
| chr11_-_83878041 | 0.07 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr22_+_50946645 | 0.07 |
ENST00000420993.2
ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2
|
non-SMC condensin II complex, subunit H2 |
| chr15_+_68346501 | 0.07 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr10_+_51565188 | 0.07 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_-_234667504 | 0.07 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr6_-_29324054 | 0.06 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr4_+_140222609 | 0.06 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr11_-_119247004 | 0.06 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_+_64681103 | 0.06 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr21_-_26797019 | 0.06 |
ENST00000440205.1
|
LINC00158
|
long intergenic non-protein coding RNA 158 |
| chr18_+_3450161 | 0.05 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_-_81046904 | 0.05 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr9_-_140142181 | 0.05 |
ENST00000484720.1
|
FAM166A
|
family with sequence similarity 166, member A |
| chr12_+_62654155 | 0.05 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr16_+_88636789 | 0.05 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr5_-_94620239 | 0.04 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr18_-_54318353 | 0.04 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr3_-_52719912 | 0.04 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr6_+_138188378 | 0.04 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_+_62654119 | 0.04 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr11_-_74660065 | 0.04 |
ENST00000525407.1
ENST00000528219.1 ENST00000531852.1 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr10_+_94608218 | 0.04 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr3_+_183899770 | 0.03 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr9_-_123639445 | 0.03 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr1_-_179198702 | 0.03 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_+_219081817 | 0.03 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr3_+_113667354 | 0.03 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr4_-_100871506 | 0.03 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr7_+_1272522 | 0.03 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr8_+_133787586 | 0.03 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr18_-_44775554 | 0.03 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr9_+_125027127 | 0.03 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr14_+_61995722 | 0.02 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr4_-_157892167 | 0.02 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr19_-_39735646 | 0.02 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr10_+_49514698 | 0.02 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr20_-_31124186 | 0.02 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr22_-_50946113 | 0.01 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr2_+_219524473 | 0.01 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr17_+_68071458 | 0.01 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_+_23244579 | 0.01 |
ENST00000452894.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr19_+_39759154 | 0.01 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr16_-_57481278 | 0.01 |
ENST00000567751.1
ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1
|
cytokine induced apoptosis inhibitor 1 |
| chr5_+_118406796 | 0.01 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr10_-_124768300 | 0.01 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr18_+_3449695 | 0.00 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_23244780 | 0.00 |
ENST00000396703.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr8_-_57123815 | 0.00 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.3 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.1 | 0.4 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.4 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:1903980 | positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.4 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.4 | GO:0050656 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |