Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
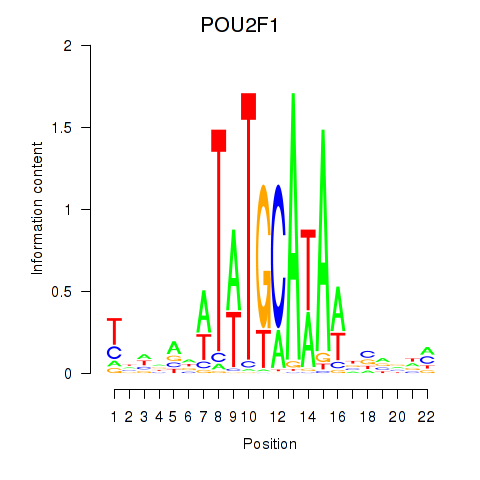
Results for POU2F1
Z-value: 2.64
Transcription factors associated with POU2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F1
|
ENSG00000143190.17 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F1 | hg19_v2_chr1_+_167298281_167298319 | -0.09 | 6.2e-01 | Click! |
Activity profile of POU2F1 motif
Sorted Z-values of POU2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_34391625 | 8.47 |
ENST00000004921.3
|
CCL18
|
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| chr14_-_106453155 | 6.87 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr14_-_106733624 | 6.69 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr2_-_238305397 | 6.62 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr14_-_106539557 | 6.09 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr14_-_106967788 | 6.00 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr21_+_10862622 | 5.87 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr2_-_89545079 | 5.81 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr2_+_89975669 | 5.61 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr2_-_89521942 | 5.57 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr2_+_89890533 | 5.54 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr15_-_22448819 | 5.52 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr14_-_106471723 | 5.24 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr14_-_107078851 | 5.14 |
ENST00000390628.2
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr14_-_106642049 | 5.13 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr12_-_111358372 | 4.92 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr22_+_23029188 | 4.84 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr22_+_23010756 | 4.79 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr2_-_89459813 | 4.64 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr16_+_33629600 | 4.58 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr2_+_89196746 | 4.48 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chr14_-_107211459 | 4.32 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr12_-_102874330 | 4.05 |
ENST00000307046.8
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr14_-_106791536 | 4.00 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_+_89999259 | 4.00 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr15_-_22473353 | 3.99 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr2_+_90060377 | 3.90 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr2_-_89292422 | 3.83 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr14_-_107283278 | 3.54 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr2_+_90458201 | 3.46 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr22_+_23089870 | 3.42 |
ENST00000390311.2
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chrX_+_15518923 | 3.26 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr2_+_36923933 | 3.11 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr11_-_101778665 | 2.93 |
ENST00000534527.1
|
ANGPTL5
|
angiopoietin-like 5 |
| chr12_-_7245125 | 2.93 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr2_+_168043793 | 2.91 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr9_-_13432977 | 2.83 |
ENST00000605459.1
|
RP11-536O18.2
|
RP11-536O18.2 |
| chr12_-_7245152 | 2.80 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr8_+_30244580 | 2.75 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr12_-_102874416 | 2.70 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr12_-_7245018 | 2.67 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr2_+_36923830 | 2.65 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr2_+_90108504 | 2.56 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr12_-_7245080 | 2.55 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr12_-_102874378 | 2.50 |
ENST00000456098.1
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr12_-_7596735 | 2.48 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr1_-_160681593 | 2.45 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr2_+_36923901 | 2.45 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr8_-_86290333 | 2.33 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr13_-_38172863 | 2.23 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr4_+_70796784 | 2.13 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr6_+_135502501 | 2.12 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_+_157154578 | 2.10 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr13_-_40924439 | 2.00 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr17_-_66951474 | 1.96 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr12_-_91546926 | 1.91 |
ENST00000550758.1
|
DCN
|
decorin |
| chr8_-_101730061 | 1.89 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_111314230 | 1.87 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr9_+_124088860 | 1.86 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr4_+_40194570 | 1.75 |
ENST00000507851.1
|
RHOH
|
ras homolog family member H |
| chr4_-_110723194 | 1.73 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr17_-_66951382 | 1.69 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr17_-_41910505 | 1.68 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr8_-_6837602 | 1.68 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr11_-_101000445 | 1.67 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr1_+_196621002 | 1.64 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr14_-_92247032 | 1.63 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr6_-_46922659 | 1.63 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr11_-_128457446 | 1.63 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr1_-_89736434 | 1.63 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr8_-_93107827 | 1.60 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_-_93107696 | 1.58 |
ENST00000436581.2
ENST00000520583.1 ENST00000519061.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_+_27849378 | 1.55 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr4_-_110723335 | 1.55 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr5_+_118668846 | 1.55 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr8_-_93107855 | 1.51 |
ENST00000517919.1
ENST00000519847.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_-_176326333 | 1.51 |
ENST00000292432.5
|
HK3
|
hexokinase 3 (white cell) |
| chr14_+_22089953 | 1.50 |
ENST00000542354.1
|
TRAV1-1
|
T cell receptor alpha variable 1-1 |
| chr9_+_112887772 | 1.49 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr4_-_152147579 | 1.47 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr4_-_110723134 | 1.47 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_-_145940214 | 1.47 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr22_-_37571089 | 1.46 |
ENST00000453962.1
ENST00000429622.1 ENST00000445595.1 |
IL2RB
|
interleukin 2 receptor, beta |
| chr2_+_204571198 | 1.43 |
ENST00000374481.3
ENST00000458610.2 ENST00000324106.8 |
CD28
|
CD28 molecule |
| chr4_+_126315091 | 1.42 |
ENST00000335110.5
|
FAT4
|
FAT atypical cadherin 4 |
| chr14_-_21270995 | 1.41 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr7_+_20686946 | 1.41 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_+_104293028 | 1.40 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr1_-_25291475 | 1.40 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr6_+_135502466 | 1.40 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr14_-_21270561 | 1.38 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr11_-_8892900 | 1.37 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr8_-_49833978 | 1.36 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr18_+_3252206 | 1.33 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_-_107729887 | 1.32 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr18_-_52989217 | 1.31 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr4_+_70861647 | 1.30 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr15_-_55541227 | 1.27 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_75230853 | 1.27 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr7_-_84121858 | 1.26 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_186570679 | 1.25 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_14693888 | 1.24 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr7_-_14026063 | 1.23 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr3_+_101546827 | 1.23 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr20_+_9146969 | 1.22 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr3_-_148939598 | 1.21 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr4_-_100065419 | 1.20 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr11_+_4673716 | 1.18 |
ENST00000530215.1
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr2_+_71205510 | 1.17 |
ENST00000272421.6
ENST00000441349.1 ENST00000457410.1 |
ANKRD53
|
ankyrin repeat domain 53 |
| chr18_+_61144160 | 1.16 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr14_+_22689792 | 1.13 |
ENST00000390462.1
|
TRAV35
|
T cell receptor alpha variable 35 |
| chr7_-_14026123 | 1.13 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr3_-_145940126 | 1.11 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr6_-_123958141 | 1.10 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr1_+_104198640 | 1.08 |
ENST00000422549.1
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr4_-_120550146 | 1.08 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr4_+_120056939 | 1.07 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr3_+_2553281 | 1.07 |
ENST00000434053.1
|
CNTN4
|
contactin 4 |
| chrX_-_6146876 | 1.07 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr13_+_24883703 | 1.06 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr6_+_135502408 | 1.05 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_-_142207004 | 1.03 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr2_+_204571375 | 1.02 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr17_+_38171681 | 1.01 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr2_+_169659121 | 1.00 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr11_-_58343319 | 0.99 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr3_+_136676851 | 0.99 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr11_+_3011093 | 0.99 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr6_-_123958051 | 0.98 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr2_-_197226875 | 0.97 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_103035102 | 0.95 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_+_169658928 | 0.91 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr1_+_62439037 | 0.89 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr6_+_116832789 | 0.87 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr17_+_38171614 | 0.86 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr6_-_123957942 | 0.85 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr6_-_123958111 | 0.84 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr2_+_71205742 | 0.82 |
ENST00000360589.3
|
ANKRD53
|
ankyrin repeat domain 53 |
| chr3_+_136676707 | 0.82 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr1_-_169703203 | 0.82 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr1_+_198608292 | 0.81 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr3_-_165555200 | 0.81 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr3_-_46069223 | 0.80 |
ENST00000309285.3
|
XCR1
|
chemokine (C motif) receptor 1 |
| chr4_-_144940477 | 0.80 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr6_-_27782548 | 0.78 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr5_-_98262240 | 0.76 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr9_-_140196703 | 0.75 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr3_-_58200398 | 0.75 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chrX_+_114827851 | 0.72 |
ENST00000539310.1
|
PLS3
|
plastin 3 |
| chr1_+_104159999 | 0.71 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr14_+_21249200 | 0.70 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr3_+_187896331 | 0.70 |
ENST00000392468.2
|
AC022498.1
|
Uncharacterized protein |
| chr17_-_41623716 | 0.70 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr8_+_18067602 | 0.69 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr11_-_107729287 | 0.69 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_+_68424434 | 0.68 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr11_-_110561721 | 0.68 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr4_+_40194609 | 0.67 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr3_-_149293990 | 0.67 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_-_107729504 | 0.67 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_173292059 | 0.65 |
ENST00000412899.1
ENST00000409532.1 |
ITGA6
|
integrin, alpha 6 |
| chr12_-_72057571 | 0.65 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr3_+_182983126 | 0.64 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr5_-_146781153 | 0.64 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr10_+_118187379 | 0.63 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr8_-_93107726 | 0.62 |
ENST00000520974.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_+_39903185 | 0.62 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_+_14294187 | 0.61 |
ENST00000573051.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr2_+_102615416 | 0.57 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr15_-_63448973 | 0.57 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr12_+_105380073 | 0.54 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chrX_-_99665262 | 0.53 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr9_+_116638562 | 0.53 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr8_-_117768023 | 0.53 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr4_-_177190364 | 0.52 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr14_-_104408645 | 0.50 |
ENST00000557640.1
|
RD3L
|
retinal degeneration 3-like |
| chr10_+_126630692 | 0.50 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chrY_+_15418467 | 0.50 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr14_+_29299437 | 0.48 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr3_+_133292574 | 0.48 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr10_+_79793518 | 0.47 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr12_-_106480587 | 0.47 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr6_+_127898312 | 0.44 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr1_+_44398943 | 0.43 |
ENST00000372359.5
ENST00000414809.3 |
ARTN
|
artemin |
| chr17_-_7387524 | 0.42 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr9_-_39239171 | 0.41 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr19_+_30156351 | 0.39 |
ENST00000592810.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr9_+_116638630 | 0.38 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr2_-_145275211 | 0.37 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_+_16500599 | 0.37 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr11_-_58980342 | 0.37 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr3_-_74570291 | 0.37 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr12_-_10601963 | 0.36 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr16_-_18462221 | 0.35 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr7_+_48211048 | 0.35 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr10_-_116286563 | 0.34 |
ENST00000369253.2
|
ABLIM1
|
actin binding LIM protein 1 |
| chr4_+_169575875 | 0.33 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_-_116286656 | 0.32 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr9_-_123812542 | 0.32 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr6_+_55192267 | 0.32 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.5 | 4.6 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.9 | 3.8 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.7 | 113.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.5 | 1.6 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.5 | 1.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 1.8 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.4 | 4.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 1.3 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.4 | 1.7 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.4 | 1.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.4 | 1.9 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.4 | 2.2 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.3 | 5.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 1.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.3 | 2.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 1.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 1.5 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 0.7 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.2 | 1.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 3.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 2.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 2.4 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.2 | 0.8 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.2 | 1.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.6 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 3.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.9 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.5 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 7.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 1.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 2.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 3.4 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.1 | 1.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.4 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 1.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 14.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 6.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.8 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 3.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 1.0 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.8 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.0 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.7 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 1.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 2.9 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 6.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.2 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 2.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.9 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 9.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 2.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 1.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.7 | 40.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.6 | 8.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 3.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 4.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 8.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 36.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.7 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.1 | 2.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 3.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 10.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 28.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 40.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.4 | 2.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.4 | 2.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 5.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 79.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.4 | 1.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.3 | 4.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 1.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 1.5 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.3 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 4.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.8 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 8.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 1.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.5 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 8.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.6 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 2.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 5.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 2.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 2.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 9.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 8.5 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 5.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 8.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 6.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 6.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.9 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 7.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 6.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 7.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 5.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |