Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
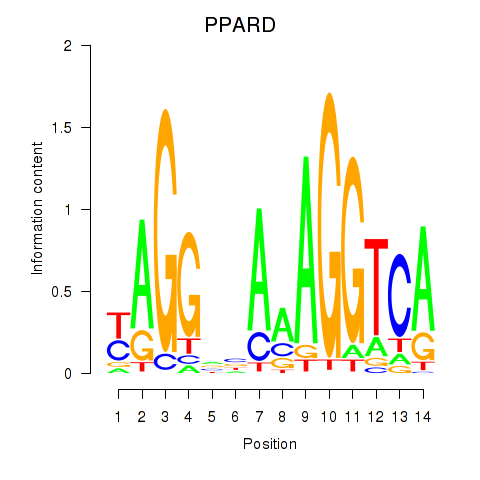
Results for PPARD
Z-value: 1.52
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310391_35310410 | -0.42 | 1.6e-02 | Click! |
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_4153598 | 8.49 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr1_-_120311517 | 6.89 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr1_-_161193349 | 6.68 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr6_+_43265992 | 6.51 |
ENST00000449231.1
ENST00000372589.3 ENST00000372585.5 |
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr6_+_43266063 | 5.41 |
ENST00000372574.3
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr10_-_74714533 | 5.23 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chr3_+_186383741 | 4.77 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr4_+_159593418 | 4.17 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr1_+_145727681 | 3.86 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr3_+_46538981 | 3.86 |
ENST00000296142.3
|
RTP3
|
receptor (chemosensory) transporter protein 3 |
| chr22_-_42526802 | 3.33 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr12_-_53320245 | 3.30 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr3_-_126236605 | 2.96 |
ENST00000290868.2
|
UROC1
|
urocanate hydratase 1 |
| chr17_+_37894179 | 2.93 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr9_-_33402506 | 2.90 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr1_-_11107280 | 2.87 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_126773880 | 2.86 |
ENST00000373615.4
|
LHX2
|
LIM homeobox 2 |
| chr1_+_6511651 | 2.84 |
ENST00000434576.1
|
ESPN
|
espin |
| chr3_-_126236588 | 2.80 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chr12_-_21757774 | 2.79 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr3_-_187009798 | 2.75 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr18_-_47340297 | 2.67 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr13_+_113760098 | 2.61 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr3_-_187009646 | 2.59 |
ENST00000296280.6
ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr20_+_42187682 | 2.50 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr6_+_32006159 | 2.22 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_+_120125245 | 2.11 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr20_+_42187608 | 2.10 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_-_28113217 | 2.07 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr7_-_87342564 | 1.97 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr6_+_32006042 | 1.96 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chrX_+_2746850 | 1.92 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr19_-_51869592 | 1.90 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr17_+_7123207 | 1.90 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr17_+_4802713 | 1.89 |
ENST00000521575.1
ENST00000381365.3 |
C17orf107
|
chromosome 17 open reading frame 107 |
| chr3_-_53878644 | 1.88 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr17_+_7123125 | 1.87 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr3_-_48936272 | 1.82 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chrX_+_2746818 | 1.80 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr17_-_34345002 | 1.76 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr17_-_34344991 | 1.61 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr4_+_140586922 | 1.57 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr9_+_115142217 | 1.47 |
ENST00000398805.3
ENST00000398803.1 ENST00000262542.7 ENST00000539114.1 |
HSDL2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_-_187009468 | 1.40 |
ENST00000425937.1
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr15_+_43885252 | 1.38 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr11_+_17281900 | 1.38 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr15_+_43985084 | 1.37 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_-_197036364 | 1.35 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr17_+_4853442 | 1.26 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_219283815 | 1.20 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr4_+_159593271 | 1.19 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr20_+_39969519 | 1.15 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chrX_+_38211777 | 1.13 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr17_+_9745786 | 1.12 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr6_+_33172407 | 1.07 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr2_+_198365095 | 1.06 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr12_-_56727676 | 1.04 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr21_-_27107881 | 1.02 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr17_+_48610042 | 1.02 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr19_-_10687983 | 1.01 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr8_-_80942467 | 0.98 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr14_+_32546274 | 0.97 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_+_48610074 | 0.93 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr14_-_51561784 | 0.92 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr8_+_145065705 | 0.89 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr12_+_121163602 | 0.82 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr7_-_34978980 | 0.79 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr12_-_56727487 | 0.79 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_-_160253998 | 0.76 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr11_-_119599794 | 0.76 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr10_-_94050820 | 0.71 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr8_-_80942061 | 0.71 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr21_-_27107344 | 0.69 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr14_+_32546145 | 0.69 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr19_+_10196981 | 0.67 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr16_-_75285380 | 0.66 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr19_+_10197463 | 0.66 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_198365122 | 0.61 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr21_-_27107198 | 0.60 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr12_+_121163538 | 0.59 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr8_-_80942139 | 0.59 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr7_+_7222157 | 0.58 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr2_-_198364552 | 0.58 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr1_-_109968973 | 0.54 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr2_-_207024134 | 0.54 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_+_115919684 | 0.53 |
ENST00000310323.8
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr2_-_198364581 | 0.52 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr21_-_27107283 | 0.52 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr19_-_49339080 | 0.46 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr17_+_73997419 | 0.45 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr9_+_35042205 | 0.43 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr8_+_145065521 | 0.39 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr19_+_29493486 | 0.37 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr4_-_7069760 | 0.36 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr9_+_131901661 | 0.34 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_62417957 | 0.33 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr9_+_131037623 | 0.33 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr19_+_29493456 | 0.32 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr17_-_4464081 | 0.31 |
ENST00000574154.1
|
GGT6
|
gamma-glutamyltransferase 6 |
| chr2_+_206547215 | 0.29 |
ENST00000360409.3
ENST00000540178.1 ENST00000540841.1 ENST00000355117.4 ENST00000450507.1 ENST00000417189.1 |
NRP2
|
neuropilin 2 |
| chr16_-_28303360 | 0.27 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr19_-_10687948 | 0.26 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr6_-_88299678 | 0.26 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr2_-_99871570 | 0.25 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr2_+_95940186 | 0.24 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr6_+_142468383 | 0.24 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr16_-_4401284 | 0.23 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr7_+_7222233 | 0.19 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr12_-_109797249 | 0.14 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr5_-_143550159 | 0.12 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr1_+_244998602 | 0.10 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr11_+_7595136 | 0.10 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr14_+_74551650 | 0.10 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr21_+_27107672 | 0.09 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr1_-_43855444 | 0.08 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr6_+_142468361 | 0.07 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr1_-_43855479 | 0.06 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr12_+_131438443 | 0.06 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr17_-_34270668 | 0.06 |
ENST00000293274.4
|
LYZL6
|
lysozyme-like 6 |
| chr1_-_11863171 | 0.05 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr6_+_88299833 | 0.05 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr12_-_95467267 | 0.02 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_+_165864821 | 0.00 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 1.2 | 4.8 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.8 | 3.3 | GO:0033076 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.8 | 5.8 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 9.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 8.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.5 | 2.6 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.5 | 11.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.5 | 1.9 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.5 | 1.8 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.4 | 2.7 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.4 | 2.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 6.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 1.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.4 | 1.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 2.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 2.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 0.7 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.9 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.2 | 11.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 4.2 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.2 | 3.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 2.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 1.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 2.9 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 2.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 3.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 5.2 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 2.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 2.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.9 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 3.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.8 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 1.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 3.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.8 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 2.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 4.6 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 1.3 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 2.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 1.1 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 6.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 5.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 4.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 4.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 3.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 16.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 9.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 2.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.8 | 5.4 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.7 | 6.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.7 | 3.7 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.7 | 2.9 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.6 | 2.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.6 | 3.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 1.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 2.0 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.5 | 3.9 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.5 | 3.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 3.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 1.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 1.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.3 | 2.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 2.0 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 1.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.3 | 2.7 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 8.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 11.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 2.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.8 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 7.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 4.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 4.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 5.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 5.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.8 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 6.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 2.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 3.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 16.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 7.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 11.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 5.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 6.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 4.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 7.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 4.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 6.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 10.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |