Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for RARA
Z-value: 1.21
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | retinoic acid receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38497640_38497647 | 0.16 | 3.8e-01 | Click! |
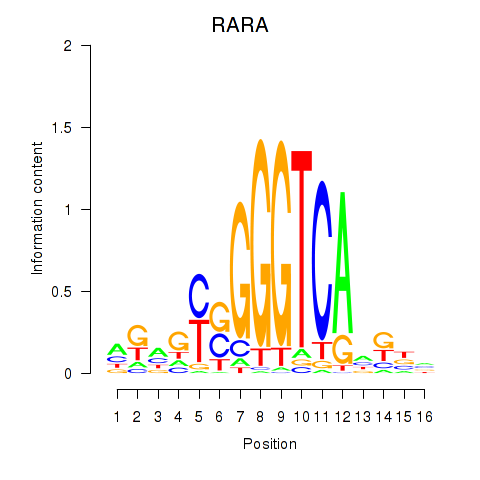
Activity profile of RARA motif
Sorted Z-values of RARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_61777459 | 2.04 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr19_-_4540486 | 1.95 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr3_+_126243126 | 1.84 |
ENST00000319340.2
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13 |
| chr9_-_116840728 | 1.78 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr7_-_100239132 | 1.78 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr3_+_108855558 | 1.74 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr12_+_54378923 | 1.68 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr4_-_48082192 | 1.65 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr17_-_61777090 | 1.57 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr10_-_52645416 | 1.55 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr15_+_33010175 | 1.50 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr11_+_45944190 | 1.44 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr3_-_112218378 | 1.37 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr22_+_45072925 | 1.36 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr1_+_145726886 | 1.36 |
ENST00000443667.1
|
PDZK1
|
PDZ domain containing 1 |
| chr14_+_103573853 | 1.34 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr3_-_112218205 | 1.31 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr2_-_235405679 | 1.21 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr1_+_6508100 | 1.18 |
ENST00000461727.1
|
ESPN
|
espin |
| chr17_+_77704681 | 1.18 |
ENST00000328313.5
|
ENPP7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr20_-_33865891 | 1.17 |
ENST00000540582.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr16_+_56995762 | 1.17 |
ENST00000200676.3
ENST00000379780.2 |
CETP
|
cholesteryl ester transfer protein, plasma |
| chr10_-_135090360 | 1.16 |
ENST00000486609.1
ENST00000445355.3 ENST00000485491.2 |
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr10_-_135238076 | 1.15 |
ENST00000414069.2
|
SPRN
|
shadow of prion protein homolog (zebrafish) |
| chr10_-_135090338 | 1.10 |
ENST00000415217.3
|
ADAM8
|
ADAM metallopeptidase domain 8 |
| chr4_-_42154895 | 1.10 |
ENST00000502486.1
ENST00000504360.1 |
BEND4
|
BEN domain containing 4 |
| chr10_-_52645379 | 1.09 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr16_-_11350036 | 1.07 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr2_-_208031542 | 1.06 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_-_8868991 | 1.03 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr6_+_36097992 | 1.03 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr3_-_49726104 | 1.01 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr22_+_45072958 | 1.00 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr20_+_44519948 | 1.00 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr1_-_156786634 | 0.99 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr12_+_54378849 | 0.98 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr22_+_50624323 | 0.97 |
ENST00000380909.4
ENST00000303434.4 |
TRABD
|
TraB domain containing |
| chr20_+_44520009 | 0.97 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chr1_-_156786530 | 0.96 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr19_+_55105085 | 0.95 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chrX_+_48534985 | 0.93 |
ENST00000450772.1
|
WAS
|
Wiskott-Aldrich syndrome |
| chr17_+_14204389 | 0.92 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr19_-_7764281 | 0.92 |
ENST00000360067.4
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr22_+_23010756 | 0.91 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr17_-_80291818 | 0.87 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr5_-_135290705 | 0.86 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr6_+_31540056 | 0.85 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr14_+_103566481 | 0.84 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr20_-_44519839 | 0.83 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr17_-_26926005 | 0.80 |
ENST00000536674.2
|
SPAG5
|
sperm associated antigen 5 |
| chr7_-_81399438 | 0.80 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_148725544 | 0.79 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr19_-_58864848 | 0.79 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr19_-_54784937 | 0.79 |
ENST00000434421.1
ENST00000314446.5 ENST00000391749.4 |
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr2_+_192542879 | 0.78 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr2_+_192543153 | 0.78 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr11_-_65626753 | 0.78 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr22_-_50970566 | 0.78 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr19_-_19739007 | 0.76 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.3 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr11_-_65626797 | 0.76 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_46144884 | 0.75 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr22_-_50970506 | 0.73 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr1_-_40367530 | 0.72 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr3_-_49726486 | 0.72 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_-_40367668 | 0.71 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr19_+_7733929 | 0.69 |
ENST00000221515.2
|
RETN
|
resistin |
| chr17_-_34308524 | 0.69 |
ENST00000293275.3
|
CCL16
|
chemokine (C-C motif) ligand 16 |
| chr19_-_58892389 | 0.69 |
ENST00000427624.2
ENST00000597582.1 |
ZNF837
|
zinc finger protein 837 |
| chr14_+_103566665 | 0.69 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr5_-_134788086 | 0.69 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr5_+_59783941 | 0.69 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr4_-_77819002 | 0.69 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr22_+_50312379 | 0.68 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr8_-_144815966 | 0.67 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chr7_+_100136811 | 0.66 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr6_+_151358048 | 0.65 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr17_-_47287928 | 0.65 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr6_+_32709119 | 0.65 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr1_+_53098862 | 0.65 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr2_+_192542850 | 0.65 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_-_62207485 | 0.64 |
ENST00000433197.3
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr14_+_24641062 | 0.63 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chrY_+_2803322 | 0.63 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr1_+_171227069 | 0.63 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr19_-_19739321 | 0.63 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr1_+_26606608 | 0.62 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr22_+_50312316 | 0.62 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr16_+_56995854 | 0.61 |
ENST00000566128.1
|
CETP
|
cholesteryl ester transfer protein, plasma |
| chr1_-_200379129 | 0.61 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr3_-_164914640 | 0.61 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr9_-_72374848 | 0.61 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr1_+_233463507 | 0.60 |
ENST00000366623.3
ENST00000366624.3 |
MLK4
|
Mitogen-activated protein kinase kinase kinase MLK4 |
| chr6_+_96025341 | 0.60 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr3_+_186288454 | 0.59 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr7_-_81399678 | 0.59 |
ENST00000412881.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_-_204463829 | 0.59 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr19_+_7701985 | 0.58 |
ENST00000595950.1
ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2
|
syntaxin binding protein 2 |
| chr1_+_25870070 | 0.58 |
ENST00000374338.4
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr2_-_208030886 | 0.57 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr16_-_11680791 | 0.57 |
ENST00000571976.1
ENST00000413364.2 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr20_+_43538756 | 0.57 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_-_81399744 | 0.56 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_+_107599267 | 0.56 |
ENST00000361318.5
ENST00000370078.1 |
PRMT6
|
protein arginine methyltransferase 6 |
| chr12_+_5541267 | 0.56 |
ENST00000423158.3
|
NTF3
|
neurotrophin 3 |
| chr19_-_4670345 | 0.56 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr14_+_38065052 | 0.55 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr11_+_33037401 | 0.55 |
ENST00000241051.3
|
DEPDC7
|
DEP domain containing 7 |
| chr4_+_3578587 | 0.55 |
ENST00000514422.1
|
LINC00955
|
long intergenic non-protein coding RNA 955 |
| chr19_+_49927006 | 0.54 |
ENST00000576655.1
|
CTD-3148I10.1
|
golgi-associated, olfactory signaling regulator |
| chr19_+_12949251 | 0.54 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr9_+_6757634 | 0.53 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_-_200379180 | 0.53 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr7_+_155090271 | 0.52 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr22_+_21996549 | 0.51 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr22_-_19165917 | 0.51 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr15_-_74495188 | 0.51 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr14_-_64194745 | 0.51 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr1_-_226187013 | 0.51 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr19_-_49926698 | 0.50 |
ENST00000270631.1
|
PTH2
|
parathyroid hormone 2 |
| chr17_+_7476136 | 0.50 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr1_-_93426998 | 0.50 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr16_-_68014732 | 0.50 |
ENST00000268793.4
|
DPEP3
|
dipeptidase 3 |
| chr16_+_69985083 | 0.49 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr14_-_107078851 | 0.49 |
ENST00000390628.2
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr11_-_58980342 | 0.49 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr1_+_160709055 | 0.48 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr12_+_125478241 | 0.48 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr20_+_43538692 | 0.48 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_+_577697 | 0.48 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr11_+_113848216 | 0.47 |
ENST00000299961.5
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
| chr7_-_81399411 | 0.47 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr20_+_3052264 | 0.46 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr2_-_74730087 | 0.46 |
ENST00000341396.2
|
LBX2
|
ladybird homeobox 2 |
| chr9_+_114393634 | 0.46 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr2_+_85132749 | 0.46 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr1_-_161039753 | 0.45 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr19_-_12405689 | 0.45 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr12_-_125348329 | 0.45 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr16_+_3115378 | 0.45 |
ENST00000529550.1
ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32
|
interleukin 32 |
| chr16_-_11680759 | 0.45 |
ENST00000571459.1
ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr19_+_16435625 | 0.45 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr16_+_29674277 | 0.44 |
ENST00000395389.2
|
SPN
|
sialophorin |
| chr22_-_29137771 | 0.44 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr19_-_1174226 | 0.44 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr22_-_29138386 | 0.44 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr10_+_102729249 | 0.44 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr11_+_74951948 | 0.43 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr17_-_77179487 | 0.41 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr16_+_8891670 | 0.41 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr14_-_22103096 | 0.41 |
ENST00000542433.1
|
OR10G2
|
olfactory receptor, family 10, subfamily G, member 2 |
| chr9_-_123691047 | 0.41 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr1_+_160051319 | 0.41 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr1_-_16939976 | 0.40 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr1_+_91966656 | 0.40 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr11_+_67033881 | 0.40 |
ENST00000308595.5
ENST00000526285.1 |
ADRBK1
|
adrenergic, beta, receptor kinase 1 |
| chr19_-_12992244 | 0.40 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr1_+_200842083 | 0.40 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr3_-_88108192 | 0.40 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr5_-_174871136 | 0.40 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr11_-_102323489 | 0.39 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr14_+_78227105 | 0.39 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr5_-_43313574 | 0.39 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr2_-_113594279 | 0.39 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr2_+_219433281 | 0.39 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr1_+_91966384 | 0.39 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr11_-_62997124 | 0.39 |
ENST00000306494.6
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr5_+_118690466 | 0.38 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr22_-_50689640 | 0.38 |
ENST00000448072.1
|
HDAC10
|
histone deacetylase 10 |
| chr1_-_109203997 | 0.38 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr17_-_16557128 | 0.38 |
ENST00000423860.2
ENST00000311331.7 ENST00000583766.1 |
ZNF624
|
zinc finger protein 624 |
| chr3_-_33138286 | 0.38 |
ENST00000416695.2
ENST00000342462.4 ENST00000399402.3 |
TMPPE
GLB1
|
transmembrane protein with metallophosphoesterase domain galactosidase, beta 1 |
| chr6_+_139349817 | 0.38 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr5_+_118691008 | 0.37 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_-_47308100 | 0.37 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr1_-_161039647 | 0.37 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr19_-_12405606 | 0.37 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr17_+_42427826 | 0.37 |
ENST00000586443.1
|
GRN
|
granulin |
| chr17_-_61523535 | 0.36 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr1_+_2487078 | 0.36 |
ENST00000426449.1
ENST00000434817.1 ENST00000435221.2 |
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr12_+_22778116 | 0.36 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr1_+_222988464 | 0.36 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr19_+_49956426 | 0.36 |
ENST00000293350.4
ENST00000540132.1 ENST00000455361.2 ENST00000433981.2 |
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr1_-_109203648 | 0.36 |
ENST00000370031.1
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr16_-_85969774 | 0.35 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr15_-_90777277 | 0.35 |
ENST00000328649.6
|
CIB1
|
calcium and integrin binding 1 (calmyrin) |
| chr4_+_39408470 | 0.35 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr11_+_63137251 | 0.34 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr19_+_49458107 | 0.34 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr12_+_57916466 | 0.34 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr17_-_61523622 | 0.34 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr13_-_21918947 | 0.34 |
ENST00000423575.1
|
LINC00539
|
LINC00539 |
| chr7_+_143078652 | 0.34 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr1_+_2487631 | 0.34 |
ENST00000409119.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr17_-_79479789 | 0.34 |
ENST00000571691.1
ENST00000571721.1 ENST00000573283.1 ENST00000575842.1 ENST00000575087.1 ENST00000570382.1 ENST00000331925.2 |
ACTG1
|
actin, gamma 1 |
| chr9_+_140100113 | 0.34 |
ENST00000371521.4
ENST00000344894.5 ENST00000427047.2 ENST00000458322.2 |
NDOR1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr22_-_32058166 | 0.33 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr22_-_21905750 | 0.33 |
ENST00000433039.1
|
RIMBP3C
|
RIMS binding protein 3C |
| chr3_-_101232019 | 0.33 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr12_+_104324112 | 0.33 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr9_+_139780942 | 0.33 |
ENST00000247668.2
ENST00000359662.3 |
TRAF2
|
TNF receptor-associated factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:2000412 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 0.5 | 1.5 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 0.9 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.4 | 0.4 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 1.8 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 0.9 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 1.8 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.3 | 1.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 1.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 0.7 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.2 | 2.0 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.2 | 1.5 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 2.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.6 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 0.6 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.2 | 0.6 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.2 | 2.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 1.4 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.5 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.2 | 2.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.5 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 2.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.7 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.5 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 1.9 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.7 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.8 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 2.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.6 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.6 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.3 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 1.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.8 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.4 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.7 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 1.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 1.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.6 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.3 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.8 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.2 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.2 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 1.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 1.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 2.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.7 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 1.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.5 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.3 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 1.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 3.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.4 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.3 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 2.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.3 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 3.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.3 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0061008 | liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0045937 | positive regulation of phosphorus metabolic process(GO:0010562) positive regulation of phosphate metabolic process(GO:0045937) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.6 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.8 | 2.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 2.0 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.3 | 2.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 2.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 1.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.4 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 2.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.2 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 1.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 2.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.0 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.7 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 3.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 1.8 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 1.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.8 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 0.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 0.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.7 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 1.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 2.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.6 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.8 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.2 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 3.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.2 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 2.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 2.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 3.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.7 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 3.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 4.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 2.0 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 3.7 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |