Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
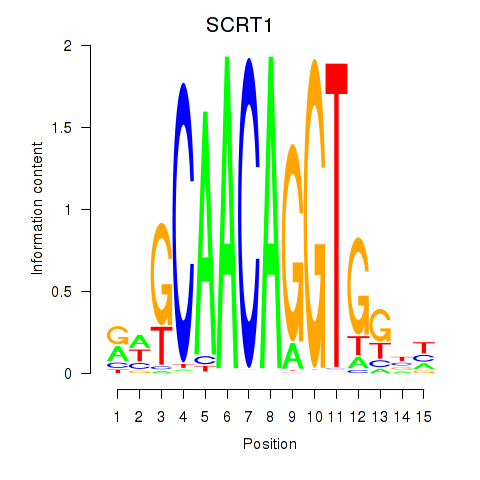
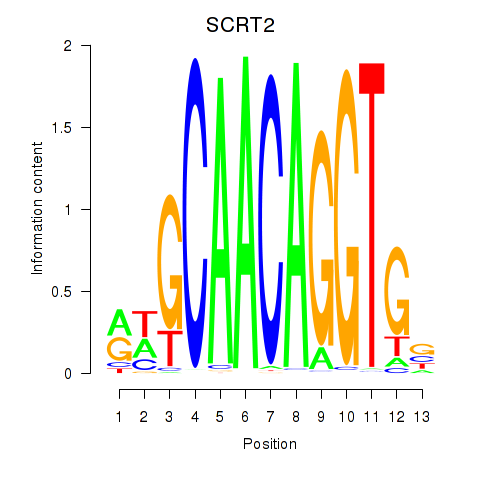
Results for SCRT1_SCRT2
Z-value: 2.17
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | 0.54 | 1.5e-03 | Click! |
| SCRT2 | hg19_v2_chr20_-_656823_656902 | 0.41 | 1.9e-02 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41258786 | 10.60 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr6_+_121756809 | 7.48 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr17_+_37784749 | 6.52 |
ENST00000394265.1
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_+_16083098 | 5.97 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_16083154 | 5.94 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr3_+_181429704 | 5.91 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr15_-_30114231 | 5.77 |
ENST00000356107.6
ENST00000545208.2 |
TJP1
|
tight junction protein 1 |
| chr3_+_16216210 | 5.55 |
ENST00000437509.1
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr1_+_16083123 | 5.43 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr7_-_122526499 | 5.35 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr4_+_41540160 | 4.94 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_15374328 | 4.92 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chrX_-_134232630 | 4.78 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr3_+_16216137 | 4.74 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr15_-_30113676 | 4.70 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr19_-_7990991 | 4.43 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr7_+_20370746 | 4.40 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr17_+_53343577 | 3.76 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr2_+_18059906 | 3.72 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr5_+_15500280 | 3.69 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr8_+_75736761 | 3.62 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr4_-_5890145 | 3.56 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr4_-_110223799 | 3.50 |
ENST00000399132.1
ENST00000399126.1 ENST00000505591.1 |
COL25A1
|
collagen, type XXV, alpha 1 |
| chr13_+_39261224 | 3.30 |
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2 |
| chrX_-_70150939 | 3.20 |
ENST00000374299.3
ENST00000298085.4 |
SLC7A3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr8_+_98881268 | 3.18 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr19_-_409134 | 3.02 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chrX_-_151143140 | 3.02 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr13_+_96743093 | 2.99 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr12_-_33049690 | 2.91 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr8_+_120220561 | 2.91 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr7_-_122526411 | 2.89 |
ENST00000449022.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr3_-_147124547 | 2.83 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr7_-_122526799 | 2.76 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr1_-_95392635 | 2.71 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr9_+_69650263 | 2.69 |
ENST00000322495.3
|
AL445665.1
|
Protein LOC100996643 |
| chr15_+_63889577 | 2.63 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr9_-_35658007 | 2.55 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr4_+_165675197 | 2.55 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr2_-_241759622 | 2.55 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr2_-_224467093 | 2.54 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr15_+_63889552 | 2.47 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr11_-_111794446 | 2.40 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr14_+_23775971 | 2.38 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr2_+_101437487 | 2.38 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_+_173600671 | 2.38 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_-_55057403 | 2.37 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chrX_-_107975917 | 2.35 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr18_+_61616510 | 2.29 |
ENST00000408945.3
|
HMSD
|
histocompatibility (minor) serpin domain containing |
| chr2_-_119916459 | 2.27 |
ENST00000272520.3
|
C1QL2
|
complement component 1, q subcomponent-like 2 |
| chr10_+_6779326 | 2.24 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr15_+_43885252 | 2.19 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_+_156308403 | 2.18 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr11_-_8739566 | 2.16 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr19_-_46974741 | 2.14 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr2_-_178753465 | 2.14 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr1_-_204135450 | 2.14 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chrX_-_99986494 | 2.13 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr1_+_156308245 | 2.12 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr18_-_10791648 | 2.02 |
ENST00000583325.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr18_+_74962505 | 2.01 |
ENST00000299727.3
|
GALR1
|
galanin receptor 1 |
| chrX_-_99987088 | 1.98 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr15_+_43985084 | 1.98 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_-_141348789 | 1.96 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr19_-_46974664 | 1.95 |
ENST00000438932.2
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr3_-_62861012 | 1.95 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr4_-_89619386 | 1.90 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr4_-_110223523 | 1.87 |
ENST00000399127.1
|
COL25A1
|
collagen, type XXV, alpha 1 |
| chrX_-_31284974 | 1.84 |
ENST00000378702.4
|
DMD
|
dystrophin |
| chr14_-_24042184 | 1.82 |
ENST00000544177.1
|
JPH4
|
junctophilin 4 |
| chr5_+_32710736 | 1.82 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_+_27062272 | 1.81 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_147211226 | 1.80 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr13_+_58206655 | 1.79 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chr5_-_35938674 | 1.78 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr12_+_129028500 | 1.78 |
ENST00000315208.8
|
TMEM132C
|
transmembrane protein 132C |
| chr12_+_81471816 | 1.76 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr2_-_110962544 | 1.75 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr12_-_71148413 | 1.74 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr17_-_32484313 | 1.73 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr11_-_117748138 | 1.72 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chrX_-_31285018 | 1.72 |
ENST00000361471.4
|
DMD
|
dystrophin |
| chr3_-_62860878 | 1.71 |
ENST00000283269.9
|
CADPS
|
Ca++-dependent secretion activator |
| chr20_-_43977055 | 1.71 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr12_-_71148357 | 1.71 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr1_+_215256467 | 1.65 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chrX_-_31285042 | 1.65 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr1_+_220701456 | 1.64 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr12_-_71003568 | 1.60 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_-_179754556 | 1.59 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr11_-_61584233 | 1.57 |
ENST00000491310.1
|
FADS1
|
fatty acid desaturase 1 |
| chr18_+_32073839 | 1.54 |
ENST00000590412.1
|
DTNA
|
dystrobrevin, alpha |
| chr3_+_133465228 | 1.54 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr8_-_139509065 | 1.51 |
ENST00000395297.1
|
FAM135B
|
family with sequence similarity 135, member B |
| chr4_-_83483395 | 1.50 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr2_+_173600565 | 1.46 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr20_-_23402028 | 1.45 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr2_+_173600514 | 1.45 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_94536898 | 1.42 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_-_224467002 | 1.41 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr15_-_76352069 | 1.39 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr13_-_25746416 | 1.34 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr1_-_94312706 | 1.33 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_180726232 | 1.30 |
ENST00000410066.1
|
ZNF385B
|
zinc finger protein 385B |
| chr14_+_32964258 | 1.29 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_24397930 | 1.29 |
ENST00000295150.3
|
FAM228A
|
family with sequence similarity 228, member A |
| chr15_-_44486632 | 1.28 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr3_-_147124307 | 1.26 |
ENST00000463250.1
|
ZIC4
|
Zic family member 4 |
| chr10_+_24528108 | 1.25 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr19_-_11373128 | 1.24 |
ENST00000294618.7
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr11_-_117747607 | 1.24 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_143567262 | 1.24 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr11_-_30038490 | 1.24 |
ENST00000328224.6
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr11_-_117747434 | 1.23 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr5_+_125759140 | 1.23 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr3_+_107096188 | 1.21 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr3_+_167453026 | 1.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr5_+_125758813 | 1.19 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_+_61583968 | 1.16 |
ENST00000517839.1
|
FADS2
|
fatty acid desaturase 2 |
| chr1_-_21044489 | 1.16 |
ENST00000247986.2
|
KIF17
|
kinesin family member 17 |
| chr5_+_125758865 | 1.16 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr3_-_79816965 | 1.15 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_+_140430979 | 1.12 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr2_-_175870085 | 1.11 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr19_-_51220176 | 1.10 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chrX_+_51636629 | 1.10 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr4_-_52904425 | 1.09 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr3_+_10857885 | 1.09 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr5_+_148651469 | 1.08 |
ENST00000515000.1
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr1_-_52456352 | 1.06 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr3_+_167453493 | 1.05 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chrX_+_55744228 | 1.05 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr19_-_49250054 | 1.04 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chrX_-_51812268 | 1.04 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr16_+_2198604 | 1.03 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr10_+_3109695 | 1.02 |
ENST00000381125.4
|
PFKP
|
phosphofructokinase, platelet |
| chr4_-_90757364 | 1.02 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_61583721 | 1.02 |
ENST00000257261.6
|
FADS2
|
fatty acid desaturase 2 |
| chr17_-_45056606 | 1.01 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr7_+_73242069 | 1.00 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr6_+_71122974 | 1.00 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr6_+_69345166 | 1.00 |
ENST00000370598.1
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr16_-_53537105 | 0.99 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr4_+_165675269 | 0.98 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr13_+_20268547 | 0.98 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr11_-_117747327 | 0.96 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr5_-_73936544 | 0.96 |
ENST00000509127.2
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr5_+_173763250 | 0.96 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr8_-_33330595 | 0.95 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr1_+_24645807 | 0.95 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_-_73936451 | 0.95 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr11_-_8816375 | 0.93 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_-_84076241 | 0.93 |
ENST00000568178.1
|
SLC38A8
|
solute carrier family 38, member 8 |
| chr1_-_162838551 | 0.93 |
ENST00000367910.1
ENST00000367912.2 ENST00000367911.2 |
C1orf110
|
chromosome 1 open reading frame 110 |
| chrX_+_151806637 | 0.92 |
ENST00000370306.2
|
GABRQ
|
gamma-aminobutyric acid (GABA) A receptor, theta |
| chr5_+_148651409 | 0.91 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr10_+_24497704 | 0.91 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr10_+_24498060 | 0.91 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr17_-_36105009 | 0.90 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr15_+_43885799 | 0.89 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr2_-_237172988 | 0.89 |
ENST00000409749.3
ENST00000430053.1 |
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr17_-_15496722 | 0.88 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr7_+_6713376 | 0.88 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr11_+_69455855 | 0.87 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr7_-_50628745 | 0.87 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr7_+_73242490 | 0.87 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr11_-_26593677 | 0.87 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_67197760 | 0.86 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr12_-_21810726 | 0.85 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr1_+_24645865 | 0.85 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr2_+_223289208 | 0.85 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr9_+_125703282 | 0.84 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr16_+_2039946 | 0.84 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr19_-_58609570 | 0.84 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr3_-_55515202 | 0.83 |
ENST00000482079.1
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr11_+_107650219 | 0.82 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr10_-_116164239 | 0.82 |
ENST00000419268.1
ENST00000304129.4 ENST00000545353.1 |
AFAP1L2
|
actin filament associated protein 1-like 2 |
| chr8_-_110988070 | 0.82 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr14_+_66578299 | 0.81 |
ENST00000554187.1
ENST00000556662.1 ENST00000556291.1 ENST00000557723.1 ENST00000557050.1 |
RP11-783L4.1
|
RP11-783L4.1 |
| chr12_+_101988627 | 0.81 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chrX_+_51928002 | 0.81 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr1_+_150245099 | 0.79 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr3_-_179754733 | 0.78 |
ENST00000472994.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chrX_-_153141302 | 0.77 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr9_+_99212403 | 0.76 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr7_+_94536514 | 0.76 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr1_-_183622442 | 0.76 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr12_-_21810765 | 0.75 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr9_+_140125209 | 0.75 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr14_+_67707826 | 0.75 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr1_-_219615984 | 0.72 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr11_-_71159380 | 0.72 |
ENST00000525346.1
ENST00000531364.1 ENST00000529990.1 ENST00000527316.1 ENST00000407721.2 |
DHCR7
|
7-dehydrocholesterol reductase |
| chr21_+_25801041 | 0.70 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr3_-_179754706 | 0.70 |
ENST00000465751.1
ENST00000467460.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr1_-_212588157 | 0.70 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr6_-_34113856 | 0.70 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr11_+_61583772 | 0.68 |
ENST00000522639.1
ENST00000522056.1 |
FADS2
|
fatty acid desaturase 2 |
| chr3_+_26664291 | 0.68 |
ENST00000396641.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr3_+_145782358 | 0.67 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr11_+_65779283 | 0.67 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.6 | 10.5 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.9 | 7.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.6 | 14.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.2 | 3.6 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 1.0 | 3.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.8 | 17.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.7 | 3.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.6 | 2.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.6 | 6.8 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.6 | 1.8 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.6 | 6.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.6 | 1.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.5 | 3.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 | 3.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.5 | 1.4 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.5 | 3.2 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.4 | 5.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 3.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 1.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.4 | 5.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 2.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.3 | 1.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 2.0 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.3 | 1.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.3 | 1.9 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.3 | 0.9 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.3 | 1.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.3 | 1.5 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 1.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 1.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 2.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 3.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.2 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 1.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.8 | GO:0060809 | chemoattraction of serotonergic neuron axon(GO:0036517) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.2 | 1.0 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.6 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 3.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 2.4 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 0.7 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.2 | 1.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 1.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.0 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.4 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.1 | 1.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 4.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.9 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.7 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.4 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.4 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 2.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 2.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 1.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.8 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 3.5 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 2.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 10.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 2.9 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 1.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 1.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.8 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 5.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 2.0 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.9 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 2.5 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 1.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 5.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 1.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.8 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 1.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 3.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) mRNA methylation(GO:0080009) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 1.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.0 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.9 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.6 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 8.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 4.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.8 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 3.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 1.0 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 4.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.8 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 5.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 17.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 1.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 2.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.2 | 1.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 13.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 3.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 1.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 5.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 3.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 3.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 17.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 8.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 10.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 7.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 6.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 15.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 2.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.3 | 6.5 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.8 | 7.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.8 | 2.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 3.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.7 | 4.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 20.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 1.6 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.5 | 1.5 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.5 | 2.9 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.5 | 1.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 3.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 1.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.4 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 3.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 1.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 2.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.3 | 2.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 10.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 0.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 0.8 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 5.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.6 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 5.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 3.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 1.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 5.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 5.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 5.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 5.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 4.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 4.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 5.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 4.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 9.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 5.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 11.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 7.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 7.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 17.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 7.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 4.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 5.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 5.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 6.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 3.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 4.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 4.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |