Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for SIX1_SIX3_SIX2
Z-value: 1.56
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
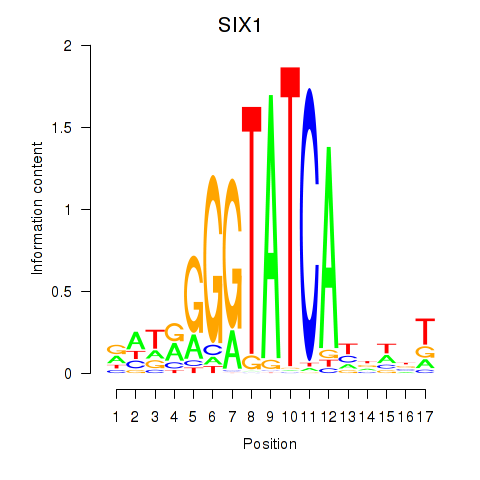
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
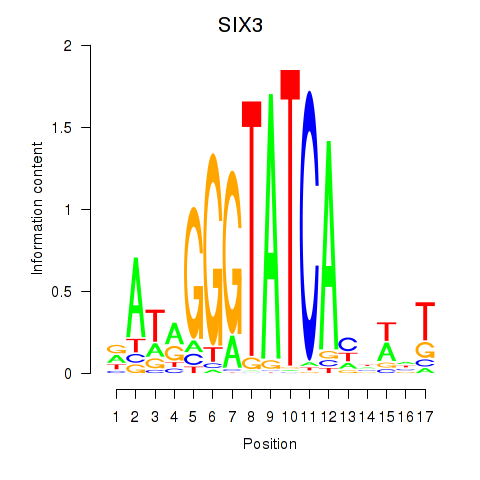
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
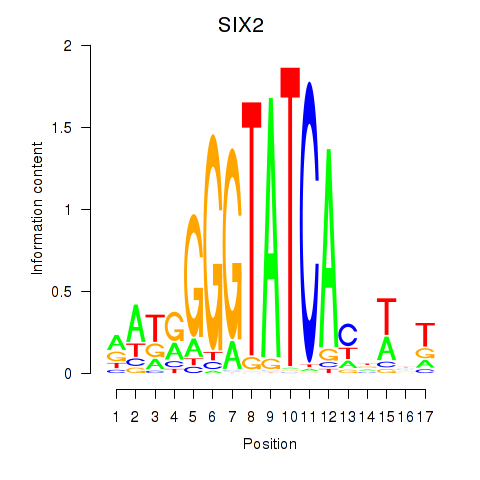
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | -0.57 | 6.4e-04 | Click! |
| SIX1 | hg19_v2_chr14_-_61116168_61116180 | -0.41 | 2.1e-02 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.04 | 8.1e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_46679144 | 6.70 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_58499434 | 6.69 |
ENST00000344743.3
ENST00000278400.3 |
GLYAT
|
glycine-N-acyltransferase |
| chr13_-_46679185 | 6.39 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_+_196946680 | 5.33 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr1_+_196946664 | 5.32 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr6_-_136847099 | 4.21 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr2_+_234826016 | 3.86 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_-_136847610 | 3.48 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr3_-_42917363 | 2.72 |
ENST00000437102.1
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr10_-_33405600 | 2.59 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr10_+_5238793 | 2.55 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr8_+_30496078 | 2.36 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr1_+_241695424 | 2.31 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_211421262 | 2.30 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chrX_+_138612889 | 2.26 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr14_+_62462541 | 2.13 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr19_+_45445491 | 2.12 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr1_+_207277590 | 2.08 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr14_-_74296806 | 2.01 |
ENST00000555539.1
|
RP5-1021I20.2
|
RP5-1021I20.2 |
| chr3_+_52448539 | 1.97 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr1_+_241695670 | 1.91 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_50836762 | 1.91 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| chr4_+_155484155 | 1.90 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr6_+_131894284 | 1.89 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr10_+_7745303 | 1.81 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr8_+_76452097 | 1.81 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr17_+_4675175 | 1.77 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr10_+_7745232 | 1.76 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_128712362 | 1.74 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr7_-_121944491 | 1.73 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr3_-_10452359 | 1.69 |
ENST00000452124.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr1_+_207277632 | 1.69 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr9_-_6645628 | 1.64 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr7_-_48068699 | 1.63 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr16_+_56666563 | 1.54 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M |
| chr1_-_12958101 | 1.41 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr12_-_55378452 | 1.40 |
ENST00000449076.1
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr5_+_56910048 | 1.36 |
ENST00000509844.1
|
CTD-2023N9.3
|
CTD-2023N9.3 |
| chr1_+_26737253 | 1.36 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr17_-_7080883 | 1.34 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr2_+_211342432 | 1.29 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr3_+_149192475 | 1.28 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_-_240906911 | 1.27 |
ENST00000431139.2
|
RP11-80B9.4
|
RP11-80B9.4 |
| chr14_+_22739823 | 1.23 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr11_-_2924720 | 1.20 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr19_-_48389651 | 1.18 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr14_-_27291313 | 1.18 |
ENST00000549330.1
|
RP11-626P14.1
|
RP11-626P14.1 |
| chr7_-_141541221 | 1.17 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr19_+_41594377 | 1.17 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr7_-_48068643 | 1.16 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr3_+_149191723 | 1.15 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr3_+_42850959 | 1.15 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr14_-_94759361 | 1.13 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_-_48068671 | 1.11 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr8_+_18248755 | 1.09 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr14_+_95047725 | 1.08 |
ENST00000554760.1
ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr5_-_35089722 | 1.06 |
ENST00000511486.1
ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR
|
prolactin receptor |
| chr11_+_27062502 | 1.04 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr8_-_29120580 | 1.04 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr9_+_6645887 | 1.03 |
ENST00000413145.1
|
RP11-390F4.6
|
RP11-390F4.6 |
| chr10_-_134331695 | 1.02 |
ENST00000455414.1
|
RP11-432J24.5
|
RP11-432J24.5 |
| chr10_+_106937525 | 0.99 |
ENST00000369699.4
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr8_+_18248786 | 0.99 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr11_+_64323428 | 0.98 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr12_-_323689 | 0.96 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr6_+_147830362 | 0.95 |
ENST00000566741.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr6_+_147981838 | 0.91 |
ENST00000427015.1
ENST00000432506.1 |
RP11-307P5.1
|
RP11-307P5.1 |
| chr3_+_113609472 | 0.90 |
ENST00000472026.1
ENST00000462838.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_-_207119738 | 0.89 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr11_+_34654011 | 0.88 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr7_-_71912046 | 0.86 |
ENST00000395276.2
ENST00000431984.1 |
CALN1
|
calneuron 1 |
| chr19_+_45445524 | 0.85 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr14_+_75761099 | 0.82 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr21_-_34185944 | 0.81 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_-_49230144 | 0.80 |
ENST00000343844.4
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr15_+_58430368 | 0.79 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr3_+_111718036 | 0.78 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr3_-_157251383 | 0.77 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_26737292 | 0.77 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr11_+_61520075 | 0.77 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr1_+_196743912 | 0.77 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr14_-_94759595 | 0.77 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_171640291 | 0.76 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chrX_+_152907913 | 0.75 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr6_+_28048753 | 0.74 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr15_+_58430567 | 0.74 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_-_125900369 | 0.73 |
ENST00000490367.1
|
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr1_+_186265399 | 0.73 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr1_+_50571949 | 0.72 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr9_+_140032842 | 0.72 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr2_+_128177253 | 0.71 |
ENST00000427769.1
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr11_+_71900572 | 0.70 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr1_+_12916941 | 0.70 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr12_-_11036844 | 0.69 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr16_-_68014732 | 0.69 |
ENST00000268793.4
|
DPEP3
|
dipeptidase 3 |
| chr19_-_56249740 | 0.69 |
ENST00000590200.1
ENST00000332836.2 |
NLRP9
|
NLR family, pyrin domain containing 9 |
| chr7_+_29519486 | 0.68 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr3_-_129147432 | 0.68 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chrX_+_144908928 | 0.67 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr8_-_29120604 | 0.66 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr3_+_111718173 | 0.65 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_+_215740709 | 0.65 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr3_+_111717600 | 0.64 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr10_+_48359344 | 0.62 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr12_+_16500037 | 0.61 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr14_+_51026844 | 0.61 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr2_-_18770802 | 0.60 |
ENST00000416783.1
|
NT5C1B
|
5'-nucleotidase, cytosolic IB |
| chr11_+_64323156 | 0.59 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr9_+_6716478 | 0.59 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr12_-_109458820 | 0.59 |
ENST00000550490.1
|
SVOP
|
SV2 related protein homolog (rat) |
| chr16_+_27214802 | 0.58 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr19_+_42055879 | 0.56 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr6_+_31554456 | 0.55 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr19_+_11457232 | 0.55 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr6_+_31554779 | 0.55 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr21_+_37442239 | 0.54 |
ENST00000530908.1
ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr22_+_36113919 | 0.53 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L, 5 |
| chr4_-_99064387 | 0.53 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr16_+_451826 | 0.52 |
ENST00000219481.5
ENST00000397710.1 ENST00000424398.2 |
DECR2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr19_+_11457175 | 0.52 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr15_+_75550940 | 0.52 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family, member C |
| chr22_+_41074754 | 0.51 |
ENST00000249016.4
|
MCHR1
|
melanin-concentrating hormone receptor 1 |
| chr1_-_169337176 | 0.51 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr14_+_58711539 | 0.51 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr6_-_159466042 | 0.51 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr6_+_88757507 | 0.49 |
ENST00000237201.1
|
SPACA1
|
sperm acrosome associated 1 |
| chr16_+_29690358 | 0.49 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr6_+_31554962 | 0.49 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr18_-_11148587 | 0.49 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr8_-_18666360 | 0.49 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_20616500 | 0.49 |
ENST00000512688.1
|
RP11-774D14.1
|
RP11-774D14.1 |
| chr16_+_56642041 | 0.48 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr17_-_45056606 | 0.47 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr21_-_33957805 | 0.47 |
ENST00000300258.3
ENST00000472557.1 |
TCP10L
|
t-complex 10-like |
| chr7_-_155326532 | 0.47 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
| chr19_+_47523058 | 0.47 |
ENST00000602212.1
ENST00000602189.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr7_-_44198874 | 0.46 |
ENST00000395796.3
ENST00000345378.2 |
GCK
|
glucokinase (hexokinase 4) |
| chr16_+_85936295 | 0.46 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr17_-_56769382 | 0.46 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr15_-_55562582 | 0.46 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr22_-_44258280 | 0.45 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr10_+_5090940 | 0.44 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr21_-_35883613 | 0.44 |
ENST00000337385.3
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr11_-_34533257 | 0.44 |
ENST00000312319.2
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr19_+_17862274 | 0.44 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr16_+_777739 | 0.43 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr12_+_56862301 | 0.43 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr20_+_18488137 | 0.43 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr10_-_43762329 | 0.43 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr22_+_32455111 | 0.42 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr17_-_34524157 | 0.42 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr7_-_142630820 | 0.42 |
ENST00000442623.1
ENST00000265310.1 |
TRPV5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr9_-_113018746 | 0.41 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr15_+_40650408 | 0.41 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr7_-_14880892 | 0.41 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_9913489 | 0.41 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr8_+_35649365 | 0.41 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr11_+_102552041 | 0.41 |
ENST00000537079.1
|
RP11-817J15.3
|
Uncharacterized protein |
| chr14_+_21214039 | 0.41 |
ENST00000326842.2
|
EDDM3A
|
epididymal protein 3A |
| chr11_+_64323098 | 0.40 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr12_+_51236703 | 0.40 |
ENST00000551456.1
ENST00000398458.3 |
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12 |
| chr1_+_53793885 | 0.40 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr1_-_157670528 | 0.39 |
ENST00000368186.5
ENST00000496769.1 |
FCRL3
|
Fc receptor-like 3 |
| chr15_+_75041184 | 0.39 |
ENST00000343932.4
|
CYP1A2
|
cytochrome P450, family 1, subfamily A, polypeptide 2 |
| chr22_-_30814469 | 0.39 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr1_-_8000872 | 0.38 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr4_+_105828492 | 0.38 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr4_-_140477910 | 0.38 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr13_-_60737898 | 0.38 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr1_-_179457805 | 0.37 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr1_+_182419261 | 0.37 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr19_+_16222439 | 0.37 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr3_-_9595480 | 0.36 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr1_+_40942887 | 0.36 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr12_-_10320256 | 0.36 |
ENST00000538745.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr4_-_48136217 | 0.36 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr6_-_167797887 | 0.36 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr17_+_73997796 | 0.36 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr1_-_57285038 | 0.35 |
ENST00000343433.6
|
C1orf168
|
chromosome 1 open reading frame 168 |
| chr20_-_1538319 | 0.35 |
ENST00000381621.1
|
SIRPD
|
signal-regulatory protein delta |
| chr3_+_101818088 | 0.35 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr7_-_34978980 | 0.35 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr5_+_140201183 | 0.34 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr11_-_108438847 | 0.34 |
ENST00000531386.1
|
EXPH5
|
exophilin 5 |
| chr11_-_15643937 | 0.34 |
ENST00000533082.1
|
RP11-531H8.2
|
RP11-531H8.2 |
| chr12_-_10321754 | 0.33 |
ENST00000539518.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr1_+_38022572 | 0.33 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr19_-_4902877 | 0.33 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr22_-_32767017 | 0.33 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr3_+_38323785 | 0.33 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr12_+_110011571 | 0.33 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr14_+_91709103 | 0.33 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr13_-_25754077 | 0.32 |
ENST00000413501.1
|
AMER2-AS1
|
AMER2 antisense RNA 1 (head to head) |
| chr7_+_150759634 | 0.32 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr5_+_125758865 | 0.32 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr3_-_19975665 | 0.32 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chrX_+_114524275 | 0.32 |
ENST00000371921.1
ENST00000451986.2 ENST00000371920.3 |
LUZP4
|
leucine zipper protein 4 |
| chr6_-_52710893 | 0.32 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr10_+_135207598 | 0.32 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr2_+_217498105 | 0.32 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.2 | 3.6 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.7 | 10.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 4.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 3.9 | GO:0050955 | thermoception(GO:0050955) |
| 0.4 | 3.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.9 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.4 | 3.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 3.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.3 | 1.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 2.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 1.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 0.5 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 6.1 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.2 | 0.7 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.2 | 1.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 0.7 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.2 | 2.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.6 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 1.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.2 | 0.4 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 1.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.7 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 1.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 2.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 2.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.2 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.5 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.2 | GO:2001202 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 2.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 6.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.0 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 1.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.5 | GO:0007167 | enzyme linked receptor protein signaling pathway(GO:0007167) |
| 0.0 | 3.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.3 | GO:0071672 | regulation of muscle hyperplasia(GO:0014738) negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.1 | GO:0032900 | viral protein processing(GO:0019082) negative regulation of neurotrophin production(GO:0032900) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) notochord regression(GO:0060032) |
| 0.0 | 0.4 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.3 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.5 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 1.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.6 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 2.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.9 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 1.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 1.8 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.3 | 1.1 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 2.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 3.0 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 1.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 2.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 7.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 3.2 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 6.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 7.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.8 | 6.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 3.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.5 | 2.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 13.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 4.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 1.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 1.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 2.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 2.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 2.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 3.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 3.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.3 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.5 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.6 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 1.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 4.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 4.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 4.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 9.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 6.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |