Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
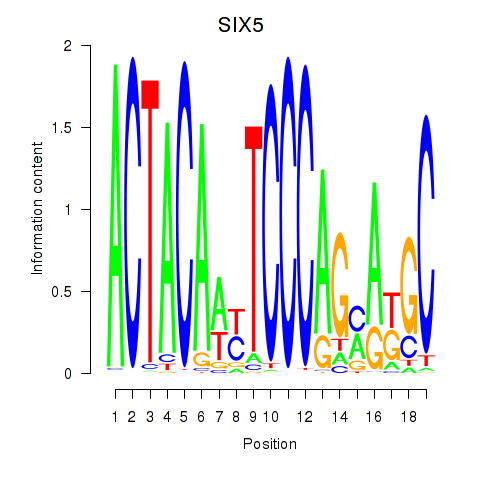
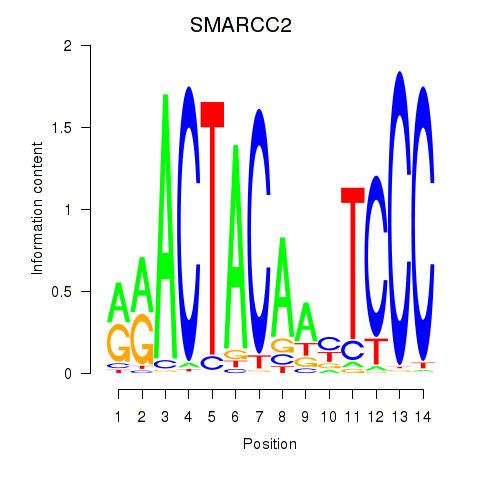
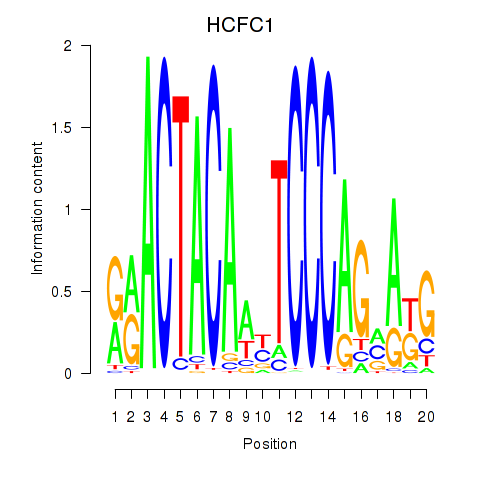
Results for SIX5_SMARCC2_HCFC1
Z-value: 4.86
Transcription factors associated with SIX5_SMARCC2_HCFC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX5
|
ENSG00000177045.6 | SIX homeobox 5 |
|
SMARCC2
|
ENSG00000139613.7 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 |
|
HCFC1
|
ENSG00000172534.9 | host cell factor C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX5 | hg19_v2_chr19_-_46272106_46272115 | -0.29 | 1.1e-01 | Click! |
| HCFC1 | hg19_v2_chrX_-_153236819_153236978 | -0.26 | 1.5e-01 | Click! |
| SMARCC2 | hg19_v2_chr12_-_56583243_56583293 | 0.03 | 8.9e-01 | Click! |
Activity profile of SIX5_SMARCC2_HCFC1 motif
Sorted Z-values of SIX5_SMARCC2_HCFC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_4791722 | 11.82 |
ENST00000269856.3
|
FEM1A
|
fem-1 homolog a (C. elegans) |
| chr19_-_51014345 | 9.03 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr1_+_45965725 | 8.77 |
ENST00000401061.4
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr19_-_51014460 | 7.64 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr4_-_8160510 | 6.58 |
ENST00000407564.3
ENST00000361737.5 ENST00000296372.8 ENST00000545242.1 ENST00000546334.1 ENST00000318888.4 ENST00000428004.2 |
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr4_-_8160416 | 6.23 |
ENST00000505872.1
ENST00000447017.2 ENST00000341937.5 ENST00000361581.5 |
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr19_-_51014588 | 5.63 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr1_-_2458026 | 5.62 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr8_-_28747424 | 5.62 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr12_+_107168418 | 5.25 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr1_+_202317855 | 5.05 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr3_-_46037299 | 4.93 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr12_+_120740119 | 4.88 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr11_-_73309112 | 4.87 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr11_-_73309228 | 4.86 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr1_-_156710916 | 4.72 |
ENST00000368211.4
|
MRPL24
|
mitochondrial ribosomal protein L24 |
| chr12_+_107168342 | 4.70 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr7_-_132766800 | 4.50 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr7_-_132766818 | 4.46 |
ENST00000262570.5
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr4_-_152682129 | 4.46 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr3_+_69788576 | 4.35 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr19_+_35225060 | 4.35 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr10_-_104262460 | 4.30 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr5_-_145562147 | 4.28 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr12_+_6961279 | 4.18 |
ENST00000229268.8
ENST00000389231.5 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr10_-_104262426 | 4.09 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr2_+_63816126 | 4.05 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr19_-_47551836 | 3.96 |
ENST00000253047.6
|
TMEM160
|
transmembrane protein 160 |
| chr20_-_62587735 | 3.93 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr1_-_156710859 | 3.93 |
ENST00000361531.2
ENST00000412846.1 |
MRPL24
|
mitochondrial ribosomal protein L24 |
| chr19_+_1241732 | 3.91 |
ENST00000215375.2
ENST00000395633.1 ENST00000591660.1 |
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr9_+_134165063 | 3.89 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr1_-_26324534 | 3.88 |
ENST00000374284.1
ENST00000441420.1 ENST00000374282.3 |
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr17_+_7358889 | 3.87 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr6_-_41703296 | 3.71 |
ENST00000373033.1
|
TFEB
|
transcription factor EB |
| chr19_-_39390350 | 3.67 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr16_+_46918235 | 3.64 |
ENST00000340124.4
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr2_+_63816269 | 3.60 |
ENST00000432309.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr13_-_41837620 | 3.53 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr6_+_42531798 | 3.52 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr14_-_73493784 | 3.52 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr16_+_2009500 | 3.44 |
ENST00000268668.6
ENST00000543683.2 ENST00000569148.1 ENST00000570172.1 |
NDUFB10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa |
| chr1_+_156024525 | 3.42 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr12_+_861717 | 3.41 |
ENST00000535572.1
|
WNK1
|
WNK lysine deficient protein kinase 1 |
| chr16_-_67840442 | 3.38 |
ENST00000536251.1
ENST00000448631.2 ENST00000602677.1 ENST00000411657.2 ENST00000425512.2 ENST00000317506.3 |
RANBP10
|
RAN binding protein 10 |
| chr12_-_54121261 | 3.33 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr9_+_134165195 | 3.33 |
ENST00000372261.1
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr17_+_67410832 | 3.32 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_-_130037162 | 3.31 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr2_+_63816295 | 3.31 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr7_+_92861653 | 3.30 |
ENST00000251739.5
ENST00000305866.5 ENST00000544910.1 ENST00000541136.1 ENST00000458530.1 ENST00000535481.1 ENST00000317751.6 |
CCDC132
|
coiled-coil domain containing 132 |
| chr5_+_175815732 | 3.29 |
ENST00000274787.2
|
HIGD2A
|
HIG1 hypoxia inducible domain family, member 2A |
| chr17_-_21117902 | 3.24 |
ENST00000317635.5
|
TMEM11
|
transmembrane protein 11 |
| chr7_-_150924121 | 3.22 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr14_-_73493825 | 3.21 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr8_+_17780346 | 3.19 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr19_-_39390440 | 3.14 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr15_-_55700457 | 3.10 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr2_+_217277137 | 3.10 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr16_+_53738053 | 3.09 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated |
| chr8_-_28747717 | 3.09 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr1_-_93645818 | 3.09 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr7_-_75988321 | 3.08 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr19_-_47354023 | 3.08 |
ENST00000601649.1
ENST00000599990.1 ENST00000352203.4 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_-_85581701 | 3.06 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr11_-_66139199 | 3.05 |
ENST00000357440.2
|
SLC29A2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr8_+_17780483 | 3.04 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr2_-_85581623 | 2.98 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr2_+_63816087 | 2.98 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr8_+_145159846 | 2.91 |
ENST00000532522.1
ENST00000527572.1 ENST00000527058.1 |
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chrX_-_130037198 | 2.89 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr10_+_60094735 | 2.87 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr14_+_36295638 | 2.85 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr19_-_47354082 | 2.83 |
ENST00000593442.1
ENST00000263270.6 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr19_-_42746714 | 2.83 |
ENST00000222330.3
|
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr13_-_30881621 | 2.82 |
ENST00000380615.3
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr12_-_54121212 | 2.82 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr18_-_43684186 | 2.76 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr19_-_5680499 | 2.74 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr1_+_156737292 | 2.70 |
ENST00000271526.4
ENST00000353233.3 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chrX_-_106362013 | 2.68 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr1_+_156024552 | 2.68 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr4_+_159593418 | 2.67 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr18_-_43684230 | 2.63 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr7_-_150754935 | 2.62 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr7_+_102389434 | 2.59 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr1_-_114355083 | 2.59 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr17_+_41322483 | 2.58 |
ENST00000341165.6
ENST00000586650.1 ENST00000422280.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr18_+_71815743 | 2.58 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr7_+_74072288 | 2.58 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr2_+_217277466 | 2.57 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr22_+_30163340 | 2.56 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr3_+_133524459 | 2.55 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr12_+_56660633 | 2.53 |
ENST00000308197.5
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr7_-_74867509 | 2.53 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr8_-_109799793 | 2.52 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr4_-_2965052 | 2.52 |
ENST00000398071.4
ENST00000502735.1 ENST00000314262.6 ENST00000416614.2 |
NOP14
|
NOP14 nucleolar protein |
| chr11_+_57425209 | 2.52 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr1_-_185126037 | 2.51 |
ENST00000367506.5
ENST00000367504.3 |
TRMT1L
|
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr9_+_116037922 | 2.51 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr1_+_44412577 | 2.50 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chrX_-_2418936 | 2.45 |
ENST00000461691.1
ENST00000381223.4 ENST00000381222.2 ENST00000412516.2 ENST00000334651.5 |
ZBED1
DHRSX
|
zinc finger, BED-type containing 1 dehydrogenase/reductase (SDR family) X-linked |
| chr19_-_39390212 | 2.45 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr1_+_1550795 | 2.41 |
ENST00000520777.1
ENST00000357210.4 ENST00000360522.4 ENST00000378710.3 ENST00000355826.5 ENST00000518681.1 ENST00000505820.2 |
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr9_-_138853156 | 2.40 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr2_-_69870747 | 2.40 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr17_+_41323204 | 2.40 |
ENST00000542611.1
ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr3_-_14166316 | 2.39 |
ENST00000396914.3
ENST00000295767.5 |
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr15_-_65477637 | 2.39 |
ENST00000300107.3
|
CLPX
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr3_-_183735651 | 2.38 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_+_2097403 | 2.38 |
ENST00000219476.3
|
TSC2
|
tuberous sclerosis 2 |
| chr15_+_55700741 | 2.38 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr19_+_35225121 | 2.37 |
ENST00000595708.1
ENST00000593781.1 |
ZNF181
|
zinc finger protein 181 |
| chr9_-_111882195 | 2.36 |
ENST00000374586.3
|
TMEM245
|
transmembrane protein 245 |
| chr19_-_5680891 | 2.36 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr13_-_41768654 | 2.35 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr10_+_124134201 | 2.35 |
ENST00000368990.3
ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr12_+_95612006 | 2.32 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_115300579 | 2.32 |
ENST00000358528.4
ENST00000525132.1 |
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr13_-_114145253 | 2.28 |
ENST00000496873.1
ENST00000478244.1 |
DCUN1D2
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr6_-_88411911 | 2.27 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr18_+_9475001 | 2.27 |
ENST00000019317.4
|
RALBP1
|
ralA binding protein 1 |
| chr19_-_5680231 | 2.25 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr6_-_41703952 | 2.25 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr1_-_144932316 | 2.23 |
ENST00000313431.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_+_95611536 | 2.23 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr15_+_41099919 | 2.21 |
ENST00000561617.1
|
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr4_+_159593271 | 2.21 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr20_-_2489542 | 2.19 |
ENST00000421216.1
ENST00000381253.1 |
ZNF343
|
zinc finger protein 343 |
| chr19_-_36980337 | 2.17 |
ENST00000434377.2
ENST00000424129.2 ENST00000452939.1 ENST00000427002.1 |
ZNF566
|
zinc finger protein 566 |
| chr11_+_134123389 | 2.16 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chr20_+_55966444 | 2.16 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr3_+_183353356 | 2.16 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr1_-_235667716 | 2.16 |
ENST00000313984.3
ENST00000366600.3 |
B3GALNT2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr7_-_135194822 | 2.13 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr10_+_74870206 | 2.13 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr1_-_115300592 | 2.12 |
ENST00000261443.5
ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr3_+_184016986 | 2.11 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr17_+_43239231 | 2.11 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_+_9102632 | 2.11 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr19_-_19249255 | 2.11 |
ENST00000587583.2
ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A
|
transmembrane protein 161A |
| chr11_-_113644491 | 2.10 |
ENST00000200135.3
|
ZW10
|
zw10 kinetochore protein |
| chr5_-_36152031 | 2.10 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr1_-_144932464 | 2.08 |
ENST00000479408.2
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_+_56661033 | 2.08 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr3_+_158991025 | 2.08 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_-_55700522 | 2.06 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr4_-_88141615 | 2.06 |
ENST00000545252.1
ENST00000425278.2 ENST00000498875.2 |
KLHL8
|
kelch-like family member 8 |
| chr1_+_1551220 | 2.05 |
ENST00000378712.1
|
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr2_-_242447732 | 2.03 |
ENST00000439101.1
ENST00000424537.1 ENST00000401869.1 ENST00000436402.1 |
STK25
|
serine/threonine kinase 25 |
| chr11_-_63933504 | 2.03 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr5_+_178487354 | 2.03 |
ENST00000315475.6
|
ZNF354C
|
zinc finger protein 354C |
| chr14_-_77923897 | 2.02 |
ENST00000343765.2
ENST00000327028.4 ENST00000556412.1 ENST00000557466.1 ENST00000448935.2 ENST00000553888.1 ENST00000557658.1 |
VIPAS39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr6_-_163148780 | 2.01 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr17_+_27046988 | 1.99 |
ENST00000496182.1
|
RPL23A
|
ribosomal protein L23a |
| chr3_-_50329990 | 1.99 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr1_-_151138422 | 1.98 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr20_-_35724388 | 1.97 |
ENST00000344359.3
ENST00000373664.3 |
RBL1
|
retinoblastoma-like 1 (p107) |
| chr15_+_75182346 | 1.97 |
ENST00000569931.1
ENST00000352410.4 ENST00000566377.1 ENST00000569233.1 ENST00000567132.1 ENST00000564633.1 ENST00000568907.1 ENST00000563422.1 ENST00000564003.1 ENST00000562800.1 ENST00000563786.1 ENST00000535694.1 ENST00000323744.6 ENST00000568828.1 ENST00000562606.1 ENST00000565576.1 ENST00000567570.1 |
MPI
|
mannose phosphate isomerase |
| chr2_-_170430366 | 1.97 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr2_-_170430277 | 1.96 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr11_-_71791518 | 1.96 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_69870835 | 1.95 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr16_-_4588822 | 1.94 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr4_-_186347099 | 1.93 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr10_+_74870253 | 1.92 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr2_+_217277271 | 1.91 |
ENST00000425815.1
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr17_+_43138664 | 1.91 |
ENST00000258960.2
|
NMT1
|
N-myristoyltransferase 1 |
| chr15_-_34635314 | 1.91 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chrX_+_49091920 | 1.91 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr10_-_121356518 | 1.91 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr17_-_5015129 | 1.91 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr5_+_85913721 | 1.90 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr1_-_151138323 | 1.89 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr10_-_94050820 | 1.89 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr15_-_55700216 | 1.88 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr3_-_43147549 | 1.88 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr13_-_30881134 | 1.88 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr19_+_58920102 | 1.87 |
ENST00000599238.1
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chr1_+_12040238 | 1.85 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr2_+_202316392 | 1.85 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr12_+_95611569 | 1.85 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_150980828 | 1.85 |
ENST00000361936.5
ENST00000361738.6 |
FAM63A
|
family with sequence similarity 63, member A |
| chr19_+_56652643 | 1.83 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr10_-_27530997 | 1.83 |
ENST00000375901.1
ENST00000412279.1 ENST00000375905.4 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_-_88141755 | 1.83 |
ENST00000273963.5
|
KLHL8
|
kelch-like family member 8 |
| chr14_+_102430855 | 1.82 |
ENST00000360184.4
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1 |
| chr19_-_36980455 | 1.82 |
ENST00000454319.1
ENST00000392170.2 |
ZNF566
|
zinc finger protein 566 |
| chr10_+_111985837 | 1.81 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr9_-_26946981 | 1.80 |
ENST00000523212.1
|
PLAA
|
phospholipase A2-activating protein |
| chr3_-_43147431 | 1.80 |
ENST00000441964.1
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr1_+_202317815 | 1.80 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_+_111985713 | 1.79 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr20_+_20033158 | 1.79 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr4_-_169931393 | 1.79 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr1_+_150980889 | 1.79 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr6_+_110501621 | 1.79 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr19_-_36980756 | 1.78 |
ENST00000493391.1
|
ZNF566
|
zinc finger protein 566 |
| chr6_-_163148700 | 1.78 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr5_+_5422778 | 1.78 |
ENST00000296564.7
|
KIAA0947
|
KIAA0947 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX5_SMARCC2_HCFC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 2.8 | 8.5 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 2.2 | 8.8 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 1.6 | 4.9 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 1.4 | 4.3 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 1.3 | 5.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 1.2 | 7.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.1 | 3.3 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 1.0 | 3.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.9 | 2.8 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.9 | 5.7 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.9 | 2.8 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.9 | 3.6 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.9 | 6.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.8 | 2.5 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.8 | 2.5 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.8 | 13.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 4.9 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.8 | 2.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.8 | 6.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.7 | 3.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.7 | 2.9 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.7 | 3.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.7 | 4.2 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.7 | 3.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.7 | 6.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 7.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 1.9 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.6 | 1.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.6 | 4.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.6 | 1.9 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 1.9 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.6 | 3.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.6 | 1.7 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.6 | 1.7 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.6 | 8.9 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.6 | 26.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 5.5 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.5 | 3.3 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.5 | 3.8 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.5 | 3.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 | 1.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.5 | 2.9 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.5 | 1.4 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.5 | 0.5 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.5 | 0.5 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.5 | 1.4 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.4 | 1.7 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.4 | 0.4 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.4 | 2.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.4 | 4.7 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.4 | 1.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.4 | 3.0 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.4 | 1.7 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.4 | 3.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 3.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 1.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.4 | 1.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 8.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 1.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.4 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 1.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.4 | 2.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.4 | 1.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.4 | 1.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.3 | 1.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.3 | 1.0 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 2.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.4 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.3 | 1.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.3 | 1.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 3.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.3 | 6.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 5.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 1.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 4.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 2.7 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.3 | 1.5 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.3 | 3.6 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 0.9 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 0.9 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.3 | 7.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 2.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.3 | 1.7 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.3 | 0.8 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.3 | 0.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.3 | 1.4 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.3 | 1.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 1.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.1 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.3 | 2.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 2.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 2.3 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.3 | 1.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 1.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 2.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 7.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 1.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 1.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 2.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.2 | 1.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.2 | 2.0 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 0.9 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 1.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 1.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 11.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 2.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 2.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 3.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 2.7 | GO:0044597 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.2 | 2.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 1.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 2.8 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.2 | 3.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 1.8 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 3.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 5.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 3.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 2.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 4.6 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.0 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 4.9 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.2 | 0.5 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 1.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.3 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.2 | 19.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 2.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 2.6 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.2 | 3.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 1.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.9 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.0 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 1.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 5.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.7 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 1.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 1.9 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 3.1 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.5 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 3.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 1.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 2.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 11.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.4 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 4.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 6.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 2.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 2.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 2.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 4.0 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 4.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.7 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.1 | 1.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:1903507 | negative regulation of nucleic acid-templated transcription(GO:1903507) |
| 0.1 | 4.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 3.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.6 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 6.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.3 | GO:1990910 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.1 | 3.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 3.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.4 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 2.9 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 0.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 1.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.4 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.3 | GO:0040009 | regulation of growth rate(GO:0040009) G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 7.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 2.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 4.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 4.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 1.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 2.9 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 2.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.1 | 1.0 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.7 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 0.9 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.7 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 12.4 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.9 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 2.9 | GO:0006363 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 6.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 2.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 3.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 3.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.7 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 4.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 2.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.8 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.9 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.3 | GO:0070142 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.1 | 0.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.6 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.0 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 1.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.2 | GO:0010628 | positive regulation of gene expression(GO:0010628) |
| 0.1 | 1.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 3.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 4.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 7.7 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 3.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 2.0 | GO:0006468 | protein phosphorylation(GO:0006468) |
| 0.0 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.4 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 3.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.5 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 5.4 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 2.1 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 12.7 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.8 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 5.3 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 1.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:1903336 | intralumenal vesicle formation(GO:0070676) negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.6 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 3.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.8 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 1.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.3 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 1.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 1.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.4 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.8 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 1.9 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 2.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 3.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 4.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.5 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 2.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 10.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.6 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 2.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 2.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 2.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 10.0 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 3.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 1.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 1.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.8 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 1.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 3.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 2.2 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:0006101 | citrate metabolic process(GO:0006101) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 3.0 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 2.4 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0070423 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 3.6 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.0 | 0.1 | GO:1903772 | virus maturation(GO:0019075) regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 2.2 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 52.2 | GO:0006351 | transcription, DNA-templated(GO:0006351) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.5 | GO:0006952 | defense response(GO:0006952) |
| 0.0 | 1.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0015813 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0051882 | mitochondrial depolarization(GO:0051882) regulation of mitochondrial depolarization(GO:0051900) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 1.3 | 10.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.3 | 6.5 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 1.0 | 4.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.9 | 9.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.9 | 4.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 2.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.7 | 10.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.7 | 5.7 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 2.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.7 | 9.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.7 | 2.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 2.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.6 | 5.5 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.6 | 3.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.5 | 8.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 2.1 | GO:0070939 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.5 | 2.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 1.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 1.3 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.4 | 2.5 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.4 | 1.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 7.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 2.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 9.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 2.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 1.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 3.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.3 | 4.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 4.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 1.7 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 1.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.3 | 1.9 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 2.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 1.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 2.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 5.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 2.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 3.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 0.7 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.2 | 5.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 9.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 2.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.9 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.2 | 0.7 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 4.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.9 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.2 | 1.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.8 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 4.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 8.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 14.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 3.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 4.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 5.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 4.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.5 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.2 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 2.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 3.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 3.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 0.5 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 5.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 14.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 5.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 3.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 3.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 12.3 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 6.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.5 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 5.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 4.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 6.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.9 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.1 | 3.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 23.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0098573 | intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.1 | 0.3 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.2 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.1 | 1.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.5 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 2.8 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 5.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 5.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 16.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 2.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 5.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 2.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 2.6 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 12.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.0 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 8.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 7.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 8.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 9.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 28.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0099568 | cytoplasmic region(GO:0099568) |
| 0.0 | 46.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 100.0 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.4 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 3.1 | 9.3 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 2.7 | 8.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 1.9 | 5.7 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 1.6 | 4.9 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.4 | 4.3 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 1.2 | 4.9 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.1 | 4.6 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 1.1 | 3.3 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 1.0 | 3.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.9 | 2.8 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.9 | 3.7 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.9 | 2.7 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.9 | 2.7 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.8 | 2.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.8 | 5.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.8 | 4.9 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.8 | 8.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.8 | 3.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.8 | 3.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.8 | 5.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.8 | 9.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 1.9 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.6 | 8.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.6 | 2.5 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 2.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.6 | 6.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.6 | 4.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.6 | 4.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.5 | 3.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 3.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.5 | 1.4 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.5 | 2.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.5 | 6.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.4 | 2.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 4.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 3.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 3.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.4 | 1.5 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.4 | 1.5 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.4 | 2.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.4 | 2.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 3.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 12.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 4.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 1.5 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.3 | 2.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 7.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.3 | 2.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.3 | 2.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 0.8 | GO:0050577 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.3 | 0.8 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.3 | 3.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 9.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 4.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 5.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 1.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 4.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 6.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.4 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 3.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 1.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 8.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 2.1 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 0.8 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 4.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 0.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 5.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 5.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 4.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 0.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 3.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 23.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 7.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 2.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 1.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 4.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 9.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 9.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 5.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 4.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 5.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 1.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 5.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 15.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.5 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 3.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 6.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 24.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 4.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 3.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 2.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.0 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 4.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 3.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 5.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 0.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 21.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 4.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 6.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.2 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.1 | 6.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 3.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 4.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 2.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.1 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 1.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 3.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 5.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.4 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 2.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 41.5 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 2.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 14.4 | GO:0003723 | RNA binding(GO:0003723) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 8.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 9.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 3.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 6.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 8.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 4.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 4.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 7.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 7.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 32.2 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.2 | 18.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 8.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 15.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 5.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 3.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 5.8 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 3.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 4.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 3.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 6.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 35.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 5.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 3.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.9 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 4.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 4.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 6.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |