Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
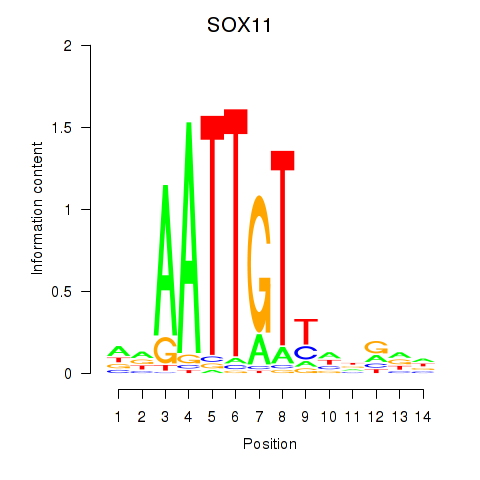
Results for SOX11
Z-value: 1.01
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | 0.02 | 9.2e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_169659121 | 3.25 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_+_169658928 | 3.01 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr12_+_41221794 | 2.39 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr17_-_66951474 | 2.35 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_+_80733570 | 2.21 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr5_+_71403280 | 2.14 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr7_-_82792215 | 2.09 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr2_+_47596287 | 2.05 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chrX_-_15619076 | 2.02 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr17_-_66951382 | 2.02 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr11_+_12399071 | 1.83 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr9_-_95166841 | 1.80 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chrX_-_13835147 | 1.78 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr2_+_102928009 | 1.59 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr2_+_166326157 | 1.59 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr12_-_22063787 | 1.56 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_+_183164574 | 1.51 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr15_-_37392086 | 1.50 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr11_+_61976137 | 1.50 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr7_-_7575477 | 1.47 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr6_-_154751629 | 1.32 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr3_+_112930306 | 1.31 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr15_+_69857515 | 1.30 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr12_+_41136144 | 1.22 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr8_-_13134045 | 1.22 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr6_-_76072719 | 1.21 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_167191814 | 1.15 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr11_-_118047376 | 1.14 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr15_-_99791413 | 1.14 |
ENST00000394129.2
ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr5_+_173472607 | 1.13 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr4_-_90759440 | 1.13 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_186570679 | 1.12 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_95972822 | 1.11 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_19796381 | 1.08 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr8_+_104831472 | 1.07 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_82165350 | 1.06 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr5_-_115872124 | 1.04 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_-_160279207 | 1.04 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr5_-_115872142 | 1.01 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_140753444 | 1.01 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr2_+_179318295 | 0.95 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr1_+_204839959 | 0.93 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr12_-_15865844 | 0.92 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr9_+_27109133 | 0.90 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr2_-_225266802 | 0.86 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr3_+_112930373 | 0.86 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr18_+_7754957 | 0.84 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr1_+_65730385 | 0.84 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_-_95657231 | 0.84 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr13_+_76413852 | 0.84 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr9_+_12775011 | 0.83 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr8_+_105352050 | 0.83 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr3_+_112930387 | 0.83 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_-_106477805 | 0.80 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr12_+_41221975 | 0.80 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr3_+_113616317 | 0.78 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr7_+_90032821 | 0.76 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr6_-_49604545 | 0.75 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr1_-_219615984 | 0.75 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr5_+_169780485 | 0.74 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr15_-_79383102 | 0.73 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr2_+_207804278 | 0.71 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr2_-_190044480 | 0.70 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_+_72204755 | 0.70 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_-_73434498 | 0.69 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr1_+_196621002 | 0.69 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr18_+_56887381 | 0.67 |
ENST00000256857.2
ENST00000529320.2 ENST00000420468.2 |
GRP
|
gastrin-releasing peptide |
| chr8_-_72274095 | 0.66 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr2_-_37068530 | 0.63 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr6_-_56492816 | 0.63 |
ENST00000522360.1
|
DST
|
dystonin |
| chr5_+_140248518 | 0.63 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr3_+_178253993 | 0.62 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr19_-_6501778 | 0.62 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr4_+_110749143 | 0.61 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr6_-_47277634 | 0.61 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr5_+_140213815 | 0.61 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr11_+_4664650 | 0.61 |
ENST00000396952.5
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr11_+_7110165 | 0.60 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr5_+_140207536 | 0.60 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr1_+_149239529 | 0.60 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr9_+_71986182 | 0.59 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_173472744 | 0.59 |
ENST00000521585.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr1_+_145525015 | 0.57 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr13_-_103019744 | 0.56 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr13_-_45048386 | 0.55 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr14_+_96949319 | 0.55 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chrX_-_32173579 | 0.54 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr4_+_159443090 | 0.54 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr9_-_114521783 | 0.54 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr1_-_24469602 | 0.53 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr7_+_15728003 | 0.53 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chr1_+_152943122 | 0.52 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr15_-_78913628 | 0.51 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr4_+_88754113 | 0.51 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_+_159442878 | 0.49 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr4_+_88754069 | 0.48 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr3_-_147124547 | 0.48 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr6_-_88001706 | 0.47 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr6_-_52705641 | 0.47 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr17_-_79895154 | 0.46 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr5_+_147582387 | 0.46 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_196621156 | 0.44 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr3_-_151160938 | 0.43 |
ENST00000489791.1
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr15_+_26147507 | 0.43 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr11_+_22694123 | 0.42 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr2_+_166430619 | 0.42 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr15_-_36544450 | 0.42 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr21_+_17792672 | 0.40 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_+_3596168 | 0.40 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr11_-_123909717 | 0.40 |
ENST00000330487.5
|
OR10G7
|
olfactory receptor, family 10, subfamily G, member 7 |
| chr7_-_112635675 | 0.40 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr17_-_79895097 | 0.39 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr1_+_154193643 | 0.39 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr2_+_138721850 | 0.39 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr7_-_92855762 | 0.38 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr8_+_24241969 | 0.38 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr2_+_105050794 | 0.38 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr19_-_14911023 | 0.38 |
ENST00000248073.2
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1 |
| chr12_-_81763127 | 0.37 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr8_+_118147498 | 0.37 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr1_-_247887345 | 0.37 |
ENST00000366485.1
|
OR14A2
|
olfactory receptor, family 14, subfamily A, member 2 |
| chr2_+_102927962 | 0.37 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr8_+_24241789 | 0.37 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr1_+_50569575 | 0.36 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_-_82373809 | 0.36 |
ENST00000379071.2
|
FABP9
|
fatty acid binding protein 9, testis |
| chr6_-_99873145 | 0.35 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chrX_+_102192200 | 0.35 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr12_+_56075330 | 0.35 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr17_+_67590125 | 0.35 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr5_-_180552304 | 0.34 |
ENST00000329365.2
|
OR2V1
|
olfactory receptor, family 2, subfamily V, member 1 |
| chr1_+_145524891 | 0.34 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr8_+_55466915 | 0.34 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr4_+_141294628 | 0.33 |
ENST00000512749.1
ENST00000608372.1 ENST00000506597.1 ENST00000394201.4 ENST00000510586.1 |
SCOC
|
short coiled-coil protein |
| chr17_-_34344991 | 0.33 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr6_-_31125850 | 0.32 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr3_+_130279178 | 0.32 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr1_-_158369256 | 0.32 |
ENST00000334438.1
|
OR10T2
|
olfactory receptor, family 10, subfamily T, member 2 |
| chr17_+_6347729 | 0.32 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chrX_+_144908928 | 0.31 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr4_-_109683691 | 0.30 |
ENST00000512320.1
ENST00000510723.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr20_-_5426332 | 0.30 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr4_+_57276661 | 0.29 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr10_-_5446786 | 0.29 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr6_-_49712123 | 0.28 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr17_+_61151306 | 0.28 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_22688150 | 0.27 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr5_-_54468974 | 0.27 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr11_-_58612168 | 0.27 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chrX_+_135388147 | 0.27 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr17_+_35732955 | 0.27 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr18_+_46065483 | 0.26 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chrM_+_7586 | 0.26 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chrX_-_100548045 | 0.26 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr6_-_49712147 | 0.25 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr17_-_34345002 | 0.25 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr13_-_40924439 | 0.25 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr12_-_79849240 | 0.25 |
ENST00000550268.1
|
RP1-78O14.1
|
RP1-78O14.1 |
| chr18_+_46065570 | 0.24 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chrX_+_27826107 | 0.24 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr17_+_35732916 | 0.24 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr4_+_144312659 | 0.23 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr9_-_21142144 | 0.23 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr1_+_214776516 | 0.22 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr14_-_20801427 | 0.22 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr10_-_24770632 | 0.22 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr12_-_11287243 | 0.22 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr4_+_159727222 | 0.21 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_+_67261008 | 0.21 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr20_+_18794370 | 0.21 |
ENST00000377428.2
|
SCP2D1
|
SCP2 sterol-binding domain containing 1 |
| chr3_-_47950745 | 0.21 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_30585486 | 0.20 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr15_+_99433570 | 0.20 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chr11_-_58611957 | 0.20 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr19_-_5680499 | 0.20 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr2_+_27799389 | 0.19 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr4_+_159443024 | 0.19 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chrX_-_132231123 | 0.19 |
ENST00000511190.1
|
USP26
|
ubiquitin specific peptidase 26 |
| chr18_+_46065393 | 0.19 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr7_+_98923505 | 0.19 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr5_+_81601166 | 0.19 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr5_+_108083517 | 0.18 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr17_-_39324424 | 0.18 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr17_-_10017864 | 0.18 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr8_+_104831554 | 0.17 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_35456444 | 0.17 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr20_-_5426380 | 0.16 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr8_+_104831440 | 0.16 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_170679024 | 0.16 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr22_-_30642728 | 0.16 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr11_+_92085262 | 0.16 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr2_-_225266711 | 0.15 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr18_+_73971121 | 0.15 |
ENST00000577797.1
|
RP11-94B19.4
|
Uncharacterized protein |
| chr5_-_86534822 | 0.15 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chrX_+_107020963 | 0.14 |
ENST00000509000.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr1_+_69091 | 0.13 |
ENST00000335137.3
|
OR4F5
|
olfactory receptor, family 4, subfamily F, member 5 |
| chr4_-_105416039 | 0.13 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr20_-_35807741 | 0.12 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr2_+_42104692 | 0.12 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr7_-_121944491 | 0.11 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr10_+_32873190 | 0.11 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 2.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 2.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 4.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 2.9 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 0.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.8 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 1.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 1.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 1.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.7 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.5 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.9 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.8 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 6.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 2.1 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 2.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 3.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 6.3 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.2 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 1.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 1.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 1.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 2.1 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 2.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 0.9 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 3.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 2.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 5.1 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 4.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 1.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.3 | 2.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 0.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 2.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 2.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 3.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 5.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 2.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |