Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for SOX17
Z-value: 2.07
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg19_v2_chr8_+_55370487_55370503 | -0.03 | 8.8e-01 | Click! |
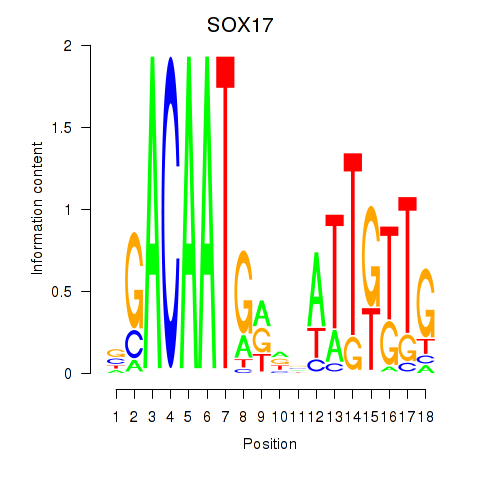
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709906 | 5.30 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr22_+_22516550 | 4.07 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr2_-_89399845 | 3.81 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr2_+_90139056 | 3.68 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr22_+_22723969 | 3.54 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr14_-_107013465 | 3.38 |
ENST00000390625.2
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr2_-_89278535 | 3.34 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr2_-_89417335 | 3.29 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr15_-_20193370 | 3.01 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr16_+_2867228 | 2.93 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr16_+_33629600 | 2.59 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr1_-_167059830 | 2.59 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr22_+_23222886 | 2.47 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr11_-_118095718 | 2.39 |
ENST00000526620.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr1_+_158901329 | 2.37 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr2_+_89196746 | 2.28 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chr22_+_42148515 | 2.12 |
ENST00000540880.1
|
MEI1
|
meiosis inhibitor 1 |
| chr19_-_19739321 | 2.11 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr12_-_127256858 | 2.07 |
ENST00000542248.1
ENST00000540684.1 |
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr14_+_22217447 | 2.05 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr14_-_106610852 | 1.99 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr14_+_22180536 | 1.99 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr14_+_22591276 | 1.98 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr2_-_90538397 | 1.97 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr9_-_35650900 | 1.96 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr15_-_81616446 | 1.92 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr1_-_160492994 | 1.92 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr5_+_54398463 | 1.91 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr11_+_60163918 | 1.91 |
ENST00000526375.1
ENST00000531783.1 ENST00000395001.1 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_+_81106951 | 1.90 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr2_+_231090471 | 1.90 |
ENST00000373645.3
|
SP140
|
SP140 nuclear body protein |
| chr6_-_25042390 | 1.87 |
ENST00000606385.1
|
RP11-367G6.3
|
RP11-367G6.3 |
| chr8_-_126963487 | 1.87 |
ENST00000518964.1
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr2_-_89327228 | 1.87 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr12_+_9822331 | 1.84 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr4_-_74864386 | 1.83 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr3_+_46412345 | 1.83 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr1_-_206306107 | 1.77 |
ENST00000436158.1
ENST00000455672.1 |
RP11-38J22.6
|
RP11-38J22.6 |
| chr11_+_60163775 | 1.77 |
ENST00000300187.6
ENST00000395005.2 |
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr8_-_126963387 | 1.77 |
ENST00000522865.1
ENST00000517869.1 |
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr14_+_22694606 | 1.76 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr2_+_89184868 | 1.75 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr2_+_90121477 | 1.74 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr7_-_121944491 | 1.74 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr14_-_107219365 | 1.74 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr7_-_81399744 | 1.72 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_-_10562356 | 1.71 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr19_-_4902877 | 1.70 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr4_-_56458374 | 1.64 |
ENST00000295645.4
|
PDCL2
|
phosducin-like 2 |
| chr14_+_22293618 | 1.61 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chr2_+_182322070 | 1.61 |
ENST00000233573.6
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr16_+_2867164 | 1.59 |
ENST00000455114.1
ENST00000450020.3 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr8_-_7309887 | 1.58 |
ENST00000458665.1
ENST00000528168.1 |
SPAG11B
|
sperm associated antigen 11B |
| chr3_-_15540055 | 1.57 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr11_-_123756334 | 1.55 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr5_+_110559312 | 1.54 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr6_-_49755019 | 1.53 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr14_+_22265444 | 1.52 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr9_+_117092149 | 1.50 |
ENST00000431067.2
ENST00000412657.1 |
ORM2
|
orosomucoid 2 |
| chrX_+_49216659 | 1.48 |
ENST00000415752.1
|
GAGE12I
|
G antigen 12I |
| chrX_+_49178536 | 1.46 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr2_+_90229045 | 1.43 |
ENST00000390278.2
|
IGKV1D-42
|
immunoglobulin kappa variable 1D-42 (non-functional) |
| chrX_-_8139308 | 1.41 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr10_-_72362515 | 1.40 |
ENST00000373209.2
ENST00000441259.1 |
PRF1
|
perforin 1 (pore forming protein) |
| chr14_+_22670455 | 1.38 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr14_+_23013015 | 1.37 |
ENST00000390535.2
|
TRAJ2
|
T cell receptor alpha joining 2 (non-functional) |
| chr2_+_232457569 | 1.34 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chrX_-_6453159 | 1.34 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chrX_+_49235708 | 1.33 |
ENST00000381725.1
|
GAGE2B
|
G antigen 2B |
| chr14_+_21236586 | 1.32 |
ENST00000326783.3
|
EDDM3B
|
epididymal protein 3B |
| chr8_-_126963417 | 1.32 |
ENST00000500989.2
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr7_+_142378566 | 1.32 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr7_+_137761167 | 1.32 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr20_-_23549334 | 1.32 |
ENST00000376979.3
|
CST9L
|
cystatin 9-like |
| chr6_+_71104588 | 1.31 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr14_-_106518922 | 1.30 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr2_+_182321925 | 1.30 |
ENST00000339307.4
ENST00000397033.2 |
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr2_-_158300556 | 1.29 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr19_-_16606988 | 1.27 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr10_+_6622345 | 1.26 |
ENST00000445427.1
ENST00000455810.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr14_-_55369525 | 1.26 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr7_-_117067541 | 1.25 |
ENST00000284629.2
|
ASZ1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr17_-_56621665 | 1.25 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr17_-_55822653 | 1.24 |
ENST00000299415.2
|
AC007431.1
|
coiled-coil domain containing 182 |
| chr3_+_111260980 | 1.22 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr10_-_127371675 | 1.21 |
ENST00000532135.1
ENST00000526819.1 ENST00000368821.3 |
TEX36
|
testis expressed 36 |
| chr1_+_47137544 | 1.21 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr1_+_207669613 | 1.21 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr11_-_116708302 | 1.19 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr22_+_44576770 | 1.18 |
ENST00000444313.3
ENST00000416291.1 |
PARVG
|
parvin, gamma |
| chr14_-_107283278 | 1.18 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr17_-_64225508 | 1.18 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_+_111260954 | 1.18 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr8_+_21155717 | 1.18 |
ENST00000522755.1
ENST00000519996.1 |
RP11-24P4.1
|
RP11-24P4.1 |
| chr1_+_84873913 | 1.17 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease II beta |
| chr12_+_127221553 | 1.16 |
ENST00000535118.1
|
LINC00943
|
long intergenic non-protein coding RNA 943 |
| chr3_-_3152031 | 1.16 |
ENST00000383846.1
ENST00000427088.1 ENST00000446632.2 ENST00000438560.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr19_+_6135646 | 1.16 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr5_+_171621176 | 1.15 |
ENST00000398186.4
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr19_+_45690646 | 1.15 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr8_+_7716700 | 1.13 |
ENST00000454911.2
ENST00000326625.5 |
SPAG11A
|
sperm associated antigen 11A |
| chr19_+_15121532 | 1.13 |
ENST00000292574.3
|
CCDC105
|
coiled-coil domain containing 105 |
| chr2_+_231090433 | 1.12 |
ENST00000486687.2
ENST00000350136.5 ENST00000392045.3 ENST00000417495.3 ENST00000343805.6 ENST00000420434.3 |
SP140
|
SP140 nuclear body protein |
| chr7_+_142494356 | 1.12 |
ENST00000390414.1
|
TRBJ2-2P
|
T cell receptor beta joining 2-2P (non-functional) |
| chr12_+_10163231 | 1.12 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr1_+_40862501 | 1.12 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr19_-_15590306 | 1.12 |
ENST00000292609.4
|
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr6_-_25042231 | 1.11 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr1_+_207669573 | 1.10 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr3_+_111260856 | 1.08 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr22_+_27053190 | 1.05 |
ENST00000439738.1
ENST00000422403.1 ENST00000436238.1 ENST00000425476.1 ENST00000455640.1 ENST00000451141.1 ENST00000452429.1 ENST00000423278.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr2_+_48844973 | 1.04 |
ENST00000403751.3
|
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr8_-_17752996 | 1.00 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr1_+_47137445 | 0.98 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chrX_+_49197607 | 0.98 |
ENST00000402590.3
|
GAGE2E
|
G antigen 2E |
| chr11_-_114430570 | 0.98 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr3_+_151451707 | 0.98 |
ENST00000356517.3
|
AADACL2
|
arylacetamide deacetylase-like 2 |
| chr3_-_123710199 | 0.95 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr1_+_174844645 | 0.94 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_+_103035102 | 0.93 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr11_+_94300474 | 0.93 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr8_-_17752912 | 0.93 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr14_+_39944025 | 0.92 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr6_+_27107053 | 0.92 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr6_-_133079022 | 0.90 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr3_+_177159695 | 0.89 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr18_-_19748379 | 0.87 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr1_+_40840320 | 0.86 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr13_-_46756351 | 0.85 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_-_111358372 | 0.85 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr14_+_22995812 | 0.84 |
ENST00000390520.1
|
TRAJ17
|
T cell receptor alpha joining 17 |
| chr11_-_118122996 | 0.84 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr2_-_220034745 | 0.84 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr12_-_498620 | 0.83 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chrY_+_27768264 | 0.83 |
ENST00000361963.2
ENST00000306609.4 |
CDY1
|
chromodomain protein, Y-linked, 1 |
| chr8_+_39442097 | 0.82 |
ENST00000265707.5
ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18
|
ADAM metallopeptidase domain 18 |
| chr19_-_16606930 | 0.82 |
ENST00000600762.1
|
CALR3
|
calreticulin 3 |
| chr7_+_137761220 | 0.82 |
ENST00000242375.3
ENST00000438242.1 |
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr2_+_149974684 | 0.81 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr13_+_25338290 | 0.81 |
ENST00000255324.5
ENST00000381921.1 ENST00000255325.6 |
RNF17
|
ring finger protein 17 |
| chrX_+_8432871 | 0.81 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr2_-_118943930 | 0.81 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr5_+_49962772 | 0.80 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr18_+_22040620 | 0.79 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr15_-_65117807 | 0.79 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr12_-_55367361 | 0.77 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr12_+_498500 | 0.77 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chrX_+_12993202 | 0.77 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr8_-_37797621 | 0.76 |
ENST00000524298.1
ENST00000307599.4 |
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr12_-_127256772 | 0.75 |
ENST00000536517.1
|
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr15_+_101459420 | 0.75 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr1_-_197744763 | 0.75 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr14_-_21056121 | 0.75 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr2_+_48844937 | 0.74 |
ENST00000448460.1
ENST00000437125.1 ENST00000430487.2 |
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr9_+_99690592 | 0.74 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr19_+_42082506 | 0.74 |
ENST00000187608.9
ENST00000401445.2 |
CEACAM21
|
carcinoembryonic antigen-related cell adhesion molecule 21 |
| chr3_+_158449972 | 0.73 |
ENST00000486568.1
|
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr12_+_9144626 | 0.73 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chrX_+_49363665 | 0.72 |
ENST00000381700.6
|
GAGE1
|
G antigen 1 |
| chr12_-_55367331 | 0.71 |
ENST00000526532.1
ENST00000532757.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr18_-_19748331 | 0.71 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr7_-_115608304 | 0.71 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr14_-_105531759 | 0.70 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr8_+_56074008 | 0.69 |
ENST00000522559.1
|
RP11-386G21.2
|
Uncharacterized protein |
| chr12_+_69742121 | 0.69 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chrX_-_13338518 | 0.68 |
ENST00000380622.2
|
ATXN3L
|
ataxin 3-like |
| chr12_+_7456880 | 0.67 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4 |
| chrY_-_19992098 | 0.67 |
ENST00000544303.1
ENST00000382867.3 |
CDY2B
|
chromodomain protein, Y-linked, 2B |
| chr1_+_207669495 | 0.67 |
ENST00000367052.1
ENST00000367051.1 ENST00000367053.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr19_+_55105085 | 0.66 |
ENST00000251372.3
ENST00000453777.1 |
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr16_-_90142338 | 0.66 |
ENST00000407825.1
ENST00000449207.2 |
PRDM7
|
PR domain containing 7 |
| chrY_+_20137667 | 0.66 |
ENST00000250838.4
ENST00000426790.1 |
CDY2A
|
chromodomain protein, Y-linked, 2A |
| chr6_-_138060103 | 0.66 |
ENST00000411615.1
ENST00000419220.1 |
RP11-356I2.1
|
RP11-356I2.1 |
| chr8_+_36641842 | 0.66 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chrX_+_134887233 | 0.66 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr4_+_2794750 | 0.65 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr6_-_52668605 | 0.65 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr2_+_10262857 | 0.64 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr11_+_18154059 | 0.62 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr6_-_26216872 | 0.62 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr19_+_41882466 | 0.62 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr11_+_6226782 | 0.61 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr11_+_60145997 | 0.61 |
ENST00000530614.1
ENST00000530027.1 ENST00000530234.2 ENST00000528215.1 ENST00000531787.1 |
MS4A7
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 7 membrane-spanning 4-domains, subfamily A, member 14 |
| chr9_+_100174232 | 0.61 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr11_-_104817919 | 0.61 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr1_-_58716197 | 0.60 |
ENST00000371234.4
|
DAB1
|
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr11_-_102496063 | 0.60 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr17_-_61959202 | 0.60 |
ENST00000449787.2
ENST00000456543.2 ENST00000423893.2 ENST00000332800.7 |
GH2
|
growth hormone 2 |
| chr8_+_27631903 | 0.60 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr18_+_9885760 | 0.60 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr1_+_182419261 | 0.60 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr12_-_68647281 | 0.58 |
ENST00000328087.4
ENST00000538666.1 |
IL22
|
interleukin 22 |
| chr8_-_7673238 | 0.58 |
ENST00000335021.2
|
DEFB107A
|
defensin, beta 107A |
| chr18_+_50278430 | 0.58 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr12_+_498545 | 0.57 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr20_-_4412320 | 0.57 |
ENST00000448825.1
|
RP11-307O10.1
|
RP11-307O10.1 |
| chr1_-_247697141 | 0.57 |
ENST00000366487.3
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3 |
| chr2_+_86668464 | 0.56 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr14_-_106370569 | 0.56 |
ENST00000390584.1
|
IGHD3-9
|
immunoglobulin heavy diversity 3-9 |
| chr18_+_20513782 | 0.56 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 1.0 | 3.0 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 1.0 | 2.9 | GO:0050904 | diapedesis(GO:0050904) |
| 0.5 | 2.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.5 | 1.5 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.4 | 1.1 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.3 | 1.3 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.3 | 1.6 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 1.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.3 | 1.2 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.3 | 1.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.6 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.3 | 0.8 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 1.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 1.4 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 1.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 1.8 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 0.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 34.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.6 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.6 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.4 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 1.9 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.4 | GO:1904730 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.1 | 1.7 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.5 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 0.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.6 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 1.4 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 1.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.8 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 1.9 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 1.9 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 2.1 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.1 | 2.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.3 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 16.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 2.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 1.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 2.0 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 5.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 3.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 5.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.8 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 1.1 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0031280 | negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0042439 | ethanolamine-containing compound metabolic process(GO:0042439) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 1.2 | GO:0007338 | single fertilization(GO:0007338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.4 | 1.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 13.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 1.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.8 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 5.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 10.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 3.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 5.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.0 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.5 | 2.7 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 1.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 1.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 1.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 2.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein antigen binding(GO:1990405) |
| 0.3 | 1.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.3 | 1.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 0.8 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.3 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 13.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.6 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.2 | 1.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 2.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 1.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.6 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 0.9 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 30.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.5 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 1.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 4.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.8 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 4.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 4.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 3.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |