Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
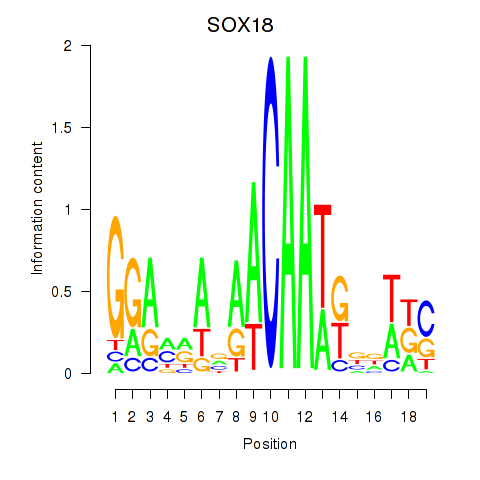
Results for SOX18
Z-value: 1.43
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | -0.25 | 1.7e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_37850026 | 2.20 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr1_-_95783809 | 2.13 |
ENST00000423410.1
|
RP4-586O15.1
|
RP4-586O15.1 |
| chr14_+_22538811 | 2.03 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr2_+_79252822 | 2.00 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr11_+_60102304 | 1.99 |
ENST00000300182.4
|
MS4A6E
|
membrane-spanning 4-domains, subfamily A, member 6E |
| chr15_+_22382382 | 1.86 |
ENST00000328795.4
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4 |
| chrX_+_30261847 | 1.72 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr5_+_127039075 | 1.71 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr2_+_79252834 | 1.69 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_+_79252804 | 1.68 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr12_+_15475462 | 1.62 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_2938044 | 1.61 |
ENST00000378404.2
|
ACTRT2
|
actin-related protein T2 |
| chr6_-_49989648 | 1.60 |
ENST00000393660.2
ENST00000371148.2 |
DEFB110
|
defensin, beta 110 locus |
| chr7_-_142120321 | 1.43 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr1_+_34326076 | 1.42 |
ENST00000519684.1
ENST00000522796.1 |
HMGB4
|
high mobility group box 4 |
| chr12_-_10562356 | 1.40 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr8_+_62737875 | 1.38 |
ENST00000523042.1
ENST00000518593.1 ENST00000519452.1 ENST00000519967.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr5_+_147648393 | 1.37 |
ENST00000511106.1
ENST00000398450.4 |
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative) |
| chr19_+_48497901 | 1.36 |
ENST00000339841.2
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr19_-_6501778 | 1.33 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr5_-_136834982 | 1.30 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr3_+_94657016 | 1.24 |
ENST00000462219.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr5_-_99870932 | 1.22 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr9_-_99064429 | 1.16 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr14_+_22392209 | 1.16 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr12_-_10573149 | 1.15 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr7_-_121944491 | 1.14 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr8_+_35649365 | 1.14 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr13_+_32313658 | 1.13 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr10_-_8095412 | 1.11 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr8_-_82443613 | 1.11 |
ENST00000360464.4
|
FABP12
|
fatty acid binding protein 12 |
| chr17_-_39041479 | 1.08 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr6_-_160679905 | 1.06 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr2_+_241544834 | 1.05 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr6_+_123317116 | 1.04 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr3_+_19189946 | 1.04 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr9_-_99064386 | 1.02 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chrY_+_3447082 | 0.96 |
ENST00000321217.4
ENST00000559055.2 |
TGIF2LY
|
TGFB-induced factor homeobox 2-like, Y-linked |
| chr5_-_101834712 | 0.93 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr17_-_8661860 | 0.92 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr7_+_10000640 | 0.91 |
ENST00000451755.1
|
AC006373.1
|
AC006373.1 |
| chr21_-_26797019 | 0.90 |
ENST00000440205.1
|
LINC00158
|
long intergenic non-protein coding RNA 158 |
| chr3_+_111717600 | 0.89 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr7_-_122339162 | 0.89 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr11_+_59807748 | 0.87 |
ENST00000278855.2
ENST00000532905.1 |
PLAC1L
|
oocyte secreted protein 2 |
| chr3_+_140947563 | 0.86 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr9_+_33240157 | 0.86 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr6_+_140175987 | 0.85 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr8_+_7397150 | 0.85 |
ENST00000533250.1
|
RP11-1118M6.1
|
proline rich 23 domain containing 1 |
| chr5_-_101834617 | 0.85 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr5_-_146461027 | 0.85 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_+_110940111 | 0.84 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr12_-_48963829 | 0.84 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr4_-_57524061 | 0.83 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr19_-_55453077 | 0.83 |
ENST00000328092.5
ENST00000590030.1 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr8_-_126963487 | 0.82 |
ENST00000518964.1
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr7_-_142176790 | 0.82 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr1_-_156828810 | 0.82 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr2_-_158345341 | 0.81 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr12_+_104697504 | 0.81 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr15_+_28624878 | 0.81 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr6_-_110500826 | 0.79 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr13_-_84456527 | 0.78 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr12_-_7904201 | 0.77 |
ENST00000354629.5
|
CLEC4C
|
C-type lectin domain family 4, member C |
| chr3_+_68055366 | 0.77 |
ENST00000496687.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chrX_-_132095419 | 0.76 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr11_+_41736067 | 0.76 |
ENST00000528720.1
|
RP11-375D13.2
|
RP11-375D13.2 |
| chr1_+_228003394 | 0.74 |
ENST00000366757.3
|
PRSS38
|
protease, serine, 38 |
| chr3_+_126911974 | 0.72 |
ENST00000398112.1
|
C3orf56
|
chromosome 3 open reading frame 56 |
| chr6_-_30899924 | 0.72 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr20_+_59654146 | 0.71 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr4_-_109541610 | 0.70 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr12_-_111127551 | 0.69 |
ENST00000439744.2
ENST00000549442.1 |
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr2_+_217277271 | 0.69 |
ENST00000425815.1
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr8_+_24151620 | 0.69 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_+_64669294 | 0.68 |
ENST00000371077.5
|
UBE2U
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr2_-_47382442 | 0.68 |
ENST00000445927.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr1_+_160051319 | 0.68 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr12_-_70093111 | 0.67 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr14_+_22217447 | 0.66 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr20_+_31499459 | 0.66 |
ENST00000439060.1
|
EFCAB8
|
EF-hand calcium binding domain 8 |
| chr8_-_7638935 | 0.66 |
ENST00000528972.1
|
AC084121.16
|
proline rich 23 domain containing 2 |
| chr5_-_43515125 | 0.66 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr12_-_70093065 | 0.65 |
ENST00000553096.1
|
BEST3
|
bestrophin 3 |
| chr11_+_73360024 | 0.65 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_+_73771844 | 0.63 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr6_-_110500905 | 0.63 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr5_-_55412774 | 0.61 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr4_-_123377880 | 0.61 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr19_-_42192189 | 0.60 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr3_+_108541545 | 0.60 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr11_+_1092184 | 0.59 |
ENST00000361558.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr12_-_70093190 | 0.59 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
| chr11_+_73358690 | 0.58 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr2_-_158345462 | 0.58 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr15_-_21071643 | 0.58 |
ENST00000454856.4
|
POTEB2
|
POTE ankyrin domain family, member B2 |
| chr12_-_8088773 | 0.58 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr15_-_22082822 | 0.57 |
ENST00000439682.1
|
POTEB
|
POTE ankyrin domain family, member B |
| chr12_+_110940005 | 0.57 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr18_+_73971121 | 0.57 |
ENST00000577797.1
|
RP11-94B19.4
|
Uncharacterized protein |
| chr1_+_209929494 | 0.56 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr3_+_35721182 | 0.54 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_+_209929377 | 0.53 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_+_73359936 | 0.53 |
ENST00000542389.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr9_+_33638033 | 0.53 |
ENST00000390389.3
|
TRBV23OR9-2
|
T cell receptor beta variable 23/OR9-2 (non-functional) |
| chrX_-_101726732 | 0.53 |
ENST00000457521.2
ENST00000412230.2 ENST00000453326.2 |
NXF2B
TCP11X2
|
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr3_-_112693759 | 0.52 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr19_-_39805976 | 0.52 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr2_-_220197351 | 0.52 |
ENST00000392083.1
|
RESP18
|
regulated endocrine-specific protein 18 |
| chr12_-_11244912 | 0.52 |
ENST00000531678.1
|
TAS2R43
|
taste receptor, type 2, member 43 |
| chr11_-_56000737 | 0.51 |
ENST00000313264.4
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2 |
| chr6_+_29427548 | 0.51 |
ENST00000377132.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chrX_+_101470280 | 0.51 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr3_+_35721106 | 0.51 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_49381572 | 0.50 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr14_+_22564294 | 0.50 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr9_+_33629119 | 0.48 |
ENST00000331828.4
|
TRBV21OR9-2
|
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr3_+_187420101 | 0.48 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr12_-_71148413 | 0.48 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr1_+_55271736 | 0.47 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr2_-_211168332 | 0.47 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr1_+_248185250 | 0.47 |
ENST00000355281.1
|
OR2L5
|
olfactory receptor, family 2, subfamily L, member 5 |
| chr1_-_153513170 | 0.47 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr17_-_28661065 | 0.47 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_-_114430169 | 0.46 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr7_+_12727250 | 0.46 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr12_+_8666126 | 0.46 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr17_+_56315787 | 0.46 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr6_-_128222103 | 0.46 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr7_-_93204033 | 0.45 |
ENST00000359558.2
ENST00000360249.4 ENST00000426151.1 |
CALCR
|
calcitonin receptor |
| chr10_-_85985294 | 0.45 |
ENST00000538192.1
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr3_-_120003941 | 0.45 |
ENST00000464295.1
|
GPR156
|
G protein-coupled receptor 156 |
| chr22_+_27017921 | 0.45 |
ENST00000354760.3
|
CRYBA4
|
crystallin, beta A4 |
| chr14_-_22938665 | 0.45 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr1_+_55446465 | 0.45 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr5_-_99870890 | 0.45 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chrX_-_110507098 | 0.45 |
ENST00000541758.1
|
CAPN6
|
calpain 6 |
| chr17_+_68071389 | 0.45 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_+_8117636 | 0.45 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr8_-_4852494 | 0.44 |
ENST00000520002.1
ENST00000602557.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr17_+_68071458 | 0.44 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_+_85430110 | 0.44 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr1_+_66258846 | 0.44 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr21_+_30671690 | 0.44 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr5_-_43515231 | 0.44 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chrX_-_70329118 | 0.43 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr19_-_43382142 | 0.43 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr3_+_160394940 | 0.43 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr4_+_109541772 | 0.43 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr11_+_123986069 | 0.42 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr9_+_137987825 | 0.42 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr8_-_94147664 | 0.41 |
ENST00000522116.1
|
C8orf87
|
chromosome 8 open reading frame 87 |
| chr12_-_75603482 | 0.41 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_+_74881355 | 0.41 |
ENST00000357877.2
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr14_+_24970800 | 0.41 |
ENST00000555109.1
|
RP11-80A15.1
|
Uncharacterized protein |
| chr3_-_112693865 | 0.41 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr19_+_4402659 | 0.41 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chrX_-_15620192 | 0.41 |
ENST00000427411.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr11_-_55371874 | 0.40 |
ENST00000302231.4
|
OR4C11
|
olfactory receptor, family 4, subfamily C, member 11 |
| chr11_-_131533462 | 0.39 |
ENST00000416725.1
|
AP003039.3
|
AP003039.3 |
| chr11_-_85397167 | 0.39 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr2_-_165811756 | 0.39 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr19_+_33571786 | 0.39 |
ENST00000170564.2
|
GPATCH1
|
G patch domain containing 1 |
| chr4_-_109541539 | 0.39 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr22_+_20850171 | 0.39 |
ENST00000445987.1
|
MED15
|
mediator complex subunit 15 |
| chr2_-_49381646 | 0.39 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr12_-_49999417 | 0.39 |
ENST00000257894.2
|
FAM186B
|
family with sequence similarity 186, member B |
| chr18_-_5396271 | 0.39 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr10_-_85985282 | 0.39 |
ENST00000372113.4
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr19_-_7040190 | 0.38 |
ENST00000381394.4
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4 |
| chr3_+_35721130 | 0.38 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_67197760 | 0.38 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_+_152483278 | 0.38 |
ENST00000334269.2
|
LCE5A
|
late cornified envelope 5A |
| chr16_-_68034470 | 0.38 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr11_+_18230685 | 0.37 |
ENST00000340135.3
ENST00000534640.1 |
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr4_+_26322987 | 0.37 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_-_105992059 | 0.37 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr11_+_20620946 | 0.37 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr12_-_68647281 | 0.37 |
ENST00000328087.4
ENST00000538666.1 |
IL22
|
interleukin 22 |
| chr1_-_155881156 | 0.35 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr12_-_71148357 | 0.35 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr5_-_78808617 | 0.35 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_+_102153859 | 0.35 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr2_-_47382414 | 0.35 |
ENST00000294947.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr6_-_27880174 | 0.35 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr6_-_33754778 | 0.34 |
ENST00000508327.1
ENST00000513701.1 |
LEMD2
|
LEM domain containing 2 |
| chr17_-_76719638 | 0.34 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr19_+_8117881 | 0.34 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr9_-_127533519 | 0.34 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr2_+_217277137 | 0.34 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr17_+_25799008 | 0.34 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr4_+_109541722 | 0.34 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chr2_+_74881398 | 0.33 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr12_-_8088871 | 0.33 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr7_+_35756066 | 0.33 |
ENST00000449644.1
|
AC018647.3
|
AC018647.3 |
| chr1_+_247768856 | 0.33 |
ENST00000320002.2
|
OR2G3
|
olfactory receptor, family 2, subfamily G, member 3 |
| chr17_+_56316054 | 0.33 |
ENST00000581008.1
|
LPO
|
lactoperoxidase |
| chr8_-_4852218 | 0.32 |
ENST00000400186.3
ENST00000602723.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr12_-_75603643 | 0.32 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr19_+_7030589 | 0.32 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.5 | 1.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 1.1 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.3 | 1.0 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.3 | 0.8 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 1.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.9 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.2 | 0.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 2.2 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 1.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 1.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.4 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 1.1 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 1.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.4 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.5 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.5 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 1.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 2.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 6.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.7 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 1.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.9 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.7 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 1.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 1.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 0.6 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.2 | 1.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.5 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 1.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.3 | 2.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 5.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 1.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 0.9 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.8 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.4 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 1.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 1.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 6.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 1.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 7.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 5.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |