Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
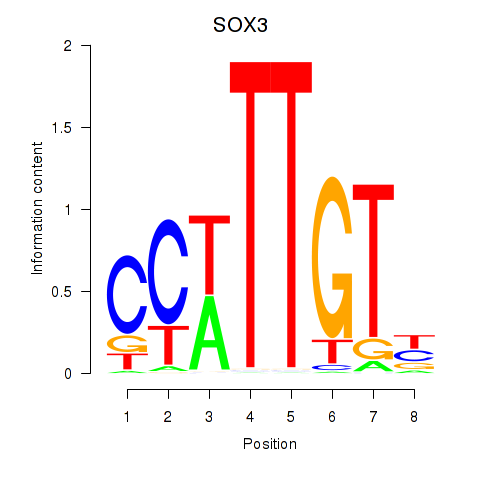
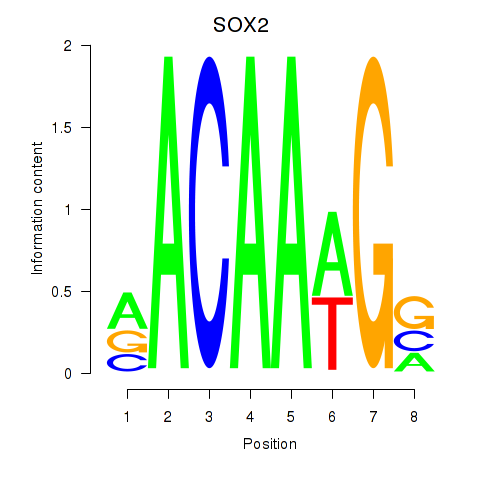
Results for SOX3_SOX2
Z-value: 3.64
Transcription factors associated with SOX3_SOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX3
|
ENSG00000134595.6 | SRY-box transcription factor 3 |
|
SOX2
|
ENSG00000181449.2 | SRY-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX2 | hg19_v2_chr3_+_181429704_181429722 | 0.72 | 3.9e-06 | Click! |
| SOX3 | hg19_v2_chrX_-_139587225_139587234 | 0.68 | 1.7e-05 | Click! |
Activity profile of SOX3_SOX2 motif
Sorted Z-values of SOX3_SOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39509070 | 10.90 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chrX_-_128788914 | 8.74 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chrX_+_103028638 | 8.73 |
ENST00000434483.1
|
PLP1
|
proteolipid protein 1 |
| chr3_+_39509163 | 8.68 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr6_-_127840336 | 8.53 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr6_-_127840021 | 8.51 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr18_-_74728998 | 7.72 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr15_+_80696666 | 7.62 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr2_-_158182410 | 7.38 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr9_+_103235365 | 7.31 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr11_-_18813353 | 7.30 |
ENST00000358540.2
ENST00000396171.4 ENST00000396167.2 |
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr5_+_94727048 | 7.22 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr2_-_158182322 | 7.18 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_+_123317116 | 7.14 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chrX_+_103029314 | 7.01 |
ENST00000429977.1
|
PLP1
|
proteolipid protein 1 |
| chr4_-_176733377 | 6.69 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr16_+_89989687 | 6.41 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr19_+_1452188 | 6.41 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr10_-_75351088 | 6.11 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr1_-_26233423 | 6.01 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr1_-_26232951 | 5.99 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr13_-_25745857 | 5.75 |
ENST00000381853.3
|
AMER2
|
APC membrane recruitment protein 2 |
| chr8_+_80523962 | 5.69 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr4_+_95916947 | 5.67 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr19_-_38720294 | 5.63 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr8_+_61969714 | 5.63 |
ENST00000522621.1
|
CLVS1
|
clavesin 1 |
| chr1_+_203096831 | 5.54 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr1_+_50571949 | 5.52 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chrX_-_139866723 | 5.51 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr4_-_5890145 | 5.47 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr20_-_52645231 | 5.47 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr5_+_140593509 | 5.43 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr10_-_128359074 | 5.37 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr20_+_42875935 | 5.29 |
ENST00000438466.1
ENST00000372952.3 ENST00000537864.1 ENST00000445952.1 |
GDAP1L1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr5_-_16936340 | 5.29 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chrX_-_24665353 | 5.19 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr5_-_146435501 | 5.18 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr18_+_21718924 | 5.13 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr5_-_114515734 | 5.12 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr18_-_31802282 | 5.11 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chrX_+_84499081 | 5.09 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr17_+_7608511 | 5.06 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr19_+_18726786 | 4.94 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr1_+_156589198 | 4.84 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr7_+_107110488 | 4.83 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chrX_+_84499038 | 4.79 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr5_-_146435694 | 4.77 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_26232522 | 4.74 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr8_-_18871159 | 4.74 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr21_+_41239243 | 4.73 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr15_+_84115868 | 4.73 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr1_+_156589051 | 4.68 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr17_-_37764128 | 4.64 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr9_+_139874683 | 4.59 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chrX_-_24665208 | 4.58 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr2_+_210444142 | 4.58 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr7_+_20370300 | 4.56 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr1_+_151682909 | 4.50 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr1_+_240255166 | 4.44 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr2_-_2334888 | 4.43 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chrX_+_84498989 | 4.39 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr9_-_73483958 | 4.38 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr8_-_21988558 | 4.37 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr11_+_60691924 | 4.36 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr1_+_150229554 | 4.35 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr7_+_20370746 | 4.31 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr8_-_22014339 | 4.28 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr19_-_38720354 | 4.27 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chrX_-_13956497 | 4.24 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr5_-_146833803 | 4.22 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_-_94614049 | 4.18 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr15_+_101402041 | 4.15 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr5_+_169931249 | 4.13 |
ENST00000520740.1
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr5_-_146435572 | 4.11 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_228337553 | 4.04 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chrX_-_138724677 | 4.03 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr7_+_154002527 | 4.03 |
ENST00000427557.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr2_+_27070964 | 4.01 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr5_-_146833485 | 3.99 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr5_+_140480083 | 3.98 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr19_+_12949251 | 3.98 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr12_-_50294033 | 3.97 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr2_+_210517895 | 3.97 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_159141397 | 3.97 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr1_+_66999268 | 3.94 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr10_+_18429606 | 3.92 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_+_162272605 | 3.92 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr2_+_210444298 | 3.92 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr20_+_20348740 | 3.88 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr8_+_105235572 | 3.84 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_26205550 | 3.82 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr17_-_56605341 | 3.81 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr2_-_99485825 | 3.81 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr2_+_17721920 | 3.80 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr2_+_27071045 | 3.80 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr6_-_127840453 | 3.80 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr9_-_91793675 | 3.80 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr14_+_101293687 | 3.76 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr8_+_85095013 | 3.73 |
ENST00000522613.1
|
RALYL
|
RALY RNA binding protein-like |
| chr2_-_158182105 | 3.72 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr19_-_821931 | 3.72 |
ENST00000359894.2
ENST00000520876.3 ENST00000519502.1 |
LPPR3
|
hsa-mir-3187 |
| chr3_+_178253993 | 3.71 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr14_-_70038032 | 3.70 |
ENST00000543986.1
|
CCDC177
|
coiled-coil domain containing 177 |
| chr2_+_210443993 | 3.68 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chrX_+_69642881 | 3.66 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr5_+_140235469 | 3.65 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr18_-_24445729 | 3.65 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr19_-_51472823 | 3.65 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr14_-_60097297 | 3.62 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr2_+_17721937 | 3.62 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr16_-_11375179 | 3.61 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr2_+_27071292 | 3.61 |
ENST00000431402.1
ENST00000434719.1 |
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr1_+_212797789 | 3.60 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr3_-_116164306 | 3.57 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_+_1411503 | 3.55 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr20_+_58203664 | 3.54 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr14_-_58618896 | 3.54 |
ENST00000267485.7
|
C14orf37
|
chromosome 14 open reading frame 37 |
| chr16_+_30934376 | 3.52 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr22_-_39052300 | 3.52 |
ENST00000355830.6
|
FAM227A
|
family with sequence similarity 227, member A |
| chrX_-_132095419 | 3.51 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_-_173638976 | 3.50 |
ENST00000333279.2
|
ANKRD45
|
ankyrin repeat domain 45 |
| chr11_-_133715394 | 3.49 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr7_+_70597109 | 3.47 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr3_+_181429704 | 3.43 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr16_+_58497567 | 3.41 |
ENST00000258187.5
|
NDRG4
|
NDRG family member 4 |
| chr2_-_175869936 | 3.39 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr17_+_36584662 | 3.33 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr18_+_21719018 | 3.32 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr4_-_87281196 | 3.30 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr17_-_9940058 | 3.30 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chrX_+_86772707 | 3.24 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr17_+_68101117 | 3.23 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr10_+_121578211 | 3.23 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chrX_+_92929192 | 3.23 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr8_-_20161466 | 3.22 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr11_+_7273181 | 3.20 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chrX_+_100333709 | 3.20 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr2_+_17721230 | 3.20 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr6_+_72596604 | 3.18 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chrX_-_138287168 | 3.18 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr5_-_176900610 | 3.14 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr2_+_155555201 | 3.14 |
ENST00000544049.1
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr10_-_128359008 | 3.11 |
ENST00000488181.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chrX_-_43832711 | 3.10 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr18_-_24445664 | 3.09 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr14_-_60097524 | 3.08 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr16_+_6069586 | 3.07 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_-_177133818 | 3.07 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr2_-_26205340 | 3.07 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr7_-_22233442 | 3.06 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr13_+_30002741 | 3.06 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr4_-_87281224 | 3.04 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_140474181 | 3.03 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr15_-_80695917 | 3.03 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr10_-_61122220 | 3.03 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chrX_+_86772787 | 3.02 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr17_-_9939935 | 3.01 |
ENST00000580043.1
|
GAS7
|
growth arrest-specific 7 |
| chr1_-_75100539 | 3.01 |
ENST00000420661.2
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr18_+_55018044 | 3.01 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr19_-_6501778 | 2.96 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr5_-_176037105 | 2.95 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr1_+_92632542 | 2.95 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chrX_+_9880412 | 2.95 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr2_+_69240302 | 2.94 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr19_-_42498369 | 2.93 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr3_-_116163830 | 2.93 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr16_+_58497587 | 2.92 |
ENST00000569404.1
ENST00000569539.1 ENST00000564126.1 ENST00000565304.1 ENST00000567667.1 |
NDRG4
|
NDRG family member 4 |
| chr19_-_42498231 | 2.90 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr2_+_69240511 | 2.89 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr8_+_30496078 | 2.88 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr6_+_69942915 | 2.87 |
ENST00000604969.1
ENST00000603207.1 |
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr8_+_79578282 | 2.87 |
ENST00000263849.4
|
ZC2HC1A
|
zinc finger, C2HC-type containing 1A |
| chrX_-_80457385 | 2.86 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_-_24690771 | 2.86 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr17_+_40119801 | 2.84 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr20_-_30458019 | 2.83 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr6_-_134639235 | 2.82 |
ENST00000533224.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr16_+_6069072 | 2.81 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr22_+_19701985 | 2.80 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr6_+_97372596 | 2.79 |
ENST00000369261.4
|
KLHL32
|
kelch-like family member 32 |
| chr1_-_31712401 | 2.79 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr1_+_50569575 | 2.78 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr10_-_104179682 | 2.77 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr9_+_95947198 | 2.77 |
ENST00000448039.1
ENST00000297954.4 ENST00000395477.2 ENST00000395475.2 ENST00000349097.3 ENST00000427277.2 ENST00000356055.3 ENST00000432730.1 |
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr7_-_112727774 | 2.77 |
ENST00000297146.3
ENST00000501255.2 |
GPR85
|
G protein-coupled receptor 85 |
| chr3_-_149688971 | 2.77 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr7_-_107968921 | 2.75 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_94537542 | 2.74 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_-_152955537 | 2.73 |
ENST00000201943.5
ENST00000539935.1 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_19813192 | 2.73 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr22_+_19702069 | 2.73 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr13_+_113656092 | 2.72 |
ENST00000397024.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr12_-_81763184 | 2.71 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr13_-_25754077 | 2.70 |
ENST00000413501.1
|
AMER2-AS1
|
AMER2 antisense RNA 1 (head to head) |
| chr11_+_66045634 | 2.70 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr10_+_73724123 | 2.70 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr4_+_150999418 | 2.70 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr1_+_200708671 | 2.69 |
ENST00000358823.2
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr16_-_75529273 | 2.68 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr15_+_84116106 | 2.68 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX3_SOX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 2.7 | 18.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.4 | 7.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.8 | 5.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 1.8 | 5.5 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 1.8 | 5.4 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 1.7 | 10.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 1.5 | 4.4 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 1.4 | 8.6 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.4 | 5.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.4 | 9.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.3 | 9.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.2 | 3.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.2 | 19.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 1.1 | 12.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 9.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.1 | 12.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.0 | 21.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.9 | 4.7 | GO:0021586 | pons maturation(GO:0021586) |
| 0.9 | 3.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 7.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.8 | 3.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.8 | 3.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.8 | 3.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.7 | 7.9 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) |
| 0.7 | 2.7 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.7 | 2.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 2.6 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.6 | 3.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 1.9 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.6 | 3.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.6 | 1.9 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.6 | 13.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.6 | 3.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.6 | 8.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.6 | 2.4 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.6 | 3.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.6 | 2.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 3.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 2.3 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.6 | 5.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.6 | 1.7 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.6 | 3.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 4.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 7.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.5 | 3.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.5 | 1.6 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.5 | 1.5 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.5 | 2.5 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.5 | 2.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.5 | 14.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 2.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.5 | 3.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.5 | 1.8 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.5 | 6.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 6.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 2.3 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.5 | 1.4 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.4 | 1.8 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.4 | 6.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 7.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.4 | 7.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 1.3 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.4 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.4 | 15.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.4 | 1.3 | GO:1905133 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.4 | 1.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.3 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.4 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 2.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.4 | 1.2 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 2.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.4 | 3.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.4 | 2.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 | 9.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 4.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.5 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.4 | 0.7 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.5 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 0.7 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.4 | 0.7 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.4 | 2.5 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.4 | 3.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 2.1 | GO:0071503 | response to heparin(GO:0071503) |
| 0.3 | 7.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.3 | 2.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.3 | 1.0 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.3 | 4.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 3.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 5.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 4.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.3 | 10.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.6 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.3 | 2.2 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.3 | 7.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 1.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.3 | 0.6 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 10.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 4.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 0.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.3 | 2.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 6.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 2.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 16.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 0.8 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 0.8 | GO:0060129 | regulation of calcium-independent cell-cell adhesion(GO:0051040) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.3 | 1.6 | GO:1901631 | postsynaptic membrane assembly(GO:0097104) positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.3 | 1.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 | 4.3 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.3 | 2.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.3 | 3.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 4.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 8.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 2.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 5.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 0.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.7 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.7 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 0.7 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 4.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 2.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 4.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 1.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 1.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 3.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 2.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.7 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 1.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 6.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 0.7 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 5.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 3.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.2 | 0.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 2.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.2 | 1.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 3.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 0.6 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.2 | 3.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 3.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 1.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 6.7 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 3.4 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.2 | 7.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 3.4 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 1.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.2 | 2.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 1.8 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 0.6 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.2 | 3.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.6 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 8.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 1.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.6 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 2.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 3.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 1.7 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.2 | 3.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.2 | 2.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 3.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 1.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 4.3 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.2 | 3.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.9 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 0.5 | GO:0032229 | negative regulation of glutamate secretion(GO:0014050) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.2 | 0.9 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 3.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.7 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.2 | 6.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.2 | 2.2 | GO:2000662 | interleukin-5 secretion(GO:0072603) regulation of interleukin-5 secretion(GO:2000662) |
| 0.2 | 2.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 1.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 1.3 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.2 | 0.5 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.2 | 0.6 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.2 | 4.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 2.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 18.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.2 | 3.7 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.8 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 2.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 2.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.6 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.8 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.1 | 32.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 2.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 5.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 3.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.8 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 1.7 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.4 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 6.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.6 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.1 | 0.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 1.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 9.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 1.0 | GO:0033007 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 1.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 9.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 1.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 1.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 2.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 3.8 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 1.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 2.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 2.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 2.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.1 | 3.4 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 4.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 11.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 3.4 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.6 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 2.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.6 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.9 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 1.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.7 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 2.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.2 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.1 | 5.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 4.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) glycosphingolipid catabolic process(GO:0046479) |
| 0.1 | 0.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 7.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.3 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.4 | GO:1901166 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 2.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:0001781 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 3.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 2.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.3 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 8.0 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 1.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.1 | 0.7 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 4.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.0 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.5 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 0.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.4 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 2.4 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.9 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.1 | GO:0071211 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 2.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 5.6 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 1.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 3.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.8 | GO:0001508 | action potential(GO:0001508) |
| 0.1 | 0.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 1.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.5 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 2.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 1.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.8 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 1.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 2.2 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 1.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0060838 | radial pattern formation(GO:0009956) lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 2.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.4 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 1.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 5.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.2 | GO:0002793 | positive regulation of peptide secretion(GO:0002793) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 2.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 4.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 1.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 3.5 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 8.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.7 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 2.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0045932 | negative regulation of muscle contraction(GO:0045932) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.4 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0014075 | response to amine(GO:0014075) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.8 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 1.6 | GO:0008652 | cellular amino acid biosynthetic process(GO:0008652) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0070570 | regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0045851 | pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 1.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.6 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.4 | 33.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.8 | 19.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.1 | 9.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.9 | 5.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.7 | 9.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 2.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.6 | 4.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.6 | 9.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.6 | 10.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 2.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 1.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 2.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.4 | 3.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 3.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.4 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 10.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 24.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 1.8 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.3 | 1.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 2.9 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.3 | 4.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.3 | 5.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 9.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 4.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 1.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 7.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 13.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 8.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 1.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 1.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.3 | 2.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 6.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 2.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 2.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 12.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 4.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 4.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.2 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 8.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 35.7 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.2 | 2.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 19.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 6.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 19.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.8 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 1.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 0.9 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 8.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 3.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 41.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 1.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 1.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.2 | 4.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 2.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 20.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 1.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 38.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.8 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 2.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 8.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 6.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 6.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.3 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.3 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 7.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 4.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 3.9 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 18.5 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 18.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 28.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 5.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 11.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 10.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 5.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 10.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 13.4 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.7 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 5.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 5.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.4 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 49.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.5 | 12.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.5 | 8.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.1 | 4.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.1 | 5.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 15.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.0 | 3.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.9 | 5.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.9 | 8.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.9 | 3.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.8 | 3.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.8 | 9.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.8 | 2.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.7 | 2.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.7 | 2.8 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.7 | 2.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.7 | 5.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.7 | 21.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.7 | 3.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.7 | 6.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.6 | 3.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.6 | 1.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.6 | 2.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.5 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.6 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.5 | 2.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.5 | 6.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 2.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 4.9 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.5 | 2.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.5 | 5.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.5 | 2.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.4 | 3.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.4 | 3.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 2.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.4 | 4.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 7.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 1.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 9.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 4.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 2.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 2.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.4 | 2.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 17.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 3.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 2.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 1.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.4 | 3.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 6.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 2.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.3 | 3.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.3 | 1.6 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.3 | 5.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 3.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.3 | 1.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 5.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 5.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 10.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 2.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 3.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 2.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 2.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.7 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 2.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 3.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.2 | 1.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 2.7 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 4.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 5.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 3.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 3.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 3.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 3.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 5.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 26.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 3.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 2.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 1.9 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.2 | 12.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 8.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 21.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 1.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 2.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 5.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 2.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 1.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.5 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 9.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.9 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 1.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 8.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 0.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 7.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.5 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.2 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 9.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.8 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.2 | 0.6 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 1.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 4.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 3.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.6 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 5.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 7.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.4 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 1.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 7.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.5 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 6.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 5.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 17.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.9 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 3.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 4.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 5.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 20.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 3.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 12.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 3.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.4 | GO:0031545 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 3.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 50.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.7 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 2.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.2 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 1.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 1.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 0.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 2.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:1904047 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 3.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 2.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 6.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 11.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 65.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 9.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 3.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 5.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 2.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 1.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 6.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 4.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 2.9 | GO:0101005 | ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 10.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 21.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 11.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 2.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 12.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 20.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 15.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 3.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 7.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 16.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 8.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 4.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 3.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.8 | 30.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 7.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 15.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 10.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 8.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 11.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 5.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 12.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.2 | 3.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 11.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 8.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 11.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 4.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 8.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 0.9 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 5.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 6.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 7.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.9 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 0.8 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 5.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 9.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 2.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 2.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 4.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 4.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 4.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |