Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
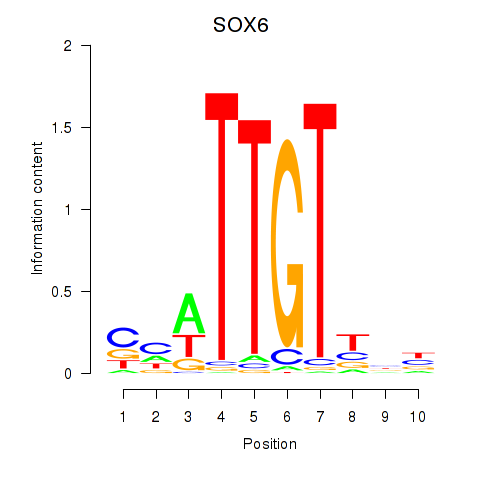
Results for SOX6
Z-value: 2.02
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16430399_16430440 | -0.40 | 2.5e-02 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_52645231 | 5.79 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_-_45270077 | 4.97 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr4_-_109090106 | 4.86 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr4_-_109089573 | 4.65 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr12_-_45269769 | 4.53 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr12_-_45270151 | 4.51 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr8_+_24151553 | 4.37 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_+_36317830 | 4.25 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr14_-_106725723 | 4.08 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr5_-_146435572 | 3.51 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_146435694 | 3.44 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_+_22748980 | 3.41 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr5_-_146435501 | 3.34 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_-_91884115 | 3.30 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr14_-_106866934 | 3.25 |
ENST00000390618.2
|
IGHV3-38
|
immunoglobulin heavy variable 3-38 (non-functional) |
| chr14_-_106845789 | 3.25 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_-_106926724 | 3.23 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr8_+_24151620 | 3.12 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_+_181429704 | 3.05 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr8_+_61822605 | 2.99 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr11_+_128563652 | 2.85 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr14_-_106518922 | 2.82 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr1_-_32801825 | 2.62 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr12_-_62586543 | 2.62 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr11_-_85430356 | 2.58 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_39509070 | 2.47 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr14_-_91884150 | 2.46 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr3_+_39509163 | 2.41 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr2_-_208031542 | 2.36 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_-_114515734 | 2.34 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr5_+_147258266 | 2.26 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr2_-_158182322 | 2.19 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_-_166702601 | 2.11 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr2_-_158182410 | 2.07 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr11_-_85430204 | 2.07 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr17_+_30813576 | 2.02 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_-_204463829 | 2.02 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_-_26233423 | 2.01 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr11_-_85430163 | 1.98 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr17_+_72667239 | 1.97 |
ENST00000402449.4
|
RAB37
|
RAB37, member RAS oncogene family |
| chrX_+_12993202 | 1.96 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr11_-_94965667 | 1.92 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr7_-_121944491 | 1.91 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr11_-_85430088 | 1.91 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_111260980 | 1.87 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr22_-_42336209 | 1.84 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr5_-_157002749 | 1.84 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr1_-_204436344 | 1.81 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr16_+_33605231 | 1.80 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr6_-_24877490 | 1.75 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr2_+_27070964 | 1.74 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr5_-_157002775 | 1.74 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr6_+_13272904 | 1.72 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_-_11548496 | 1.69 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr2_-_9143786 | 1.69 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr6_-_49712072 | 1.65 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr14_-_106791536 | 1.64 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_-_2334888 | 1.62 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr2_-_135805008 | 1.60 |
ENST00000414343.1
|
MAP3K19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr6_-_133055815 | 1.57 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr12_+_11081828 | 1.56 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr1_-_26232951 | 1.55 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr2_+_27071045 | 1.54 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr12_-_50294033 | 1.53 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr1_-_26232522 | 1.52 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr20_-_17511962 | 1.51 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr13_+_51913819 | 1.46 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr7_+_87563557 | 1.44 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr9_-_130712995 | 1.43 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr7_+_87563458 | 1.43 |
ENST00000398204.4
|
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr9_+_139874683 | 1.42 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chrX_+_84499038 | 1.42 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr12_-_11036844 | 1.42 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr6_-_49712091 | 1.39 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chrX_+_84498989 | 1.38 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr1_+_240255166 | 1.37 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr2_-_158182105 | 1.37 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr5_-_169725231 | 1.35 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr6_-_127840021 | 1.35 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr5_+_49962772 | 1.34 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_13925098 | 1.33 |
ENST00000488300.1
ENST00000544682.1 ENST00000420478.2 |
RNF182
|
ring finger protein 182 |
| chr15_-_52043722 | 1.33 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr11_-_123756334 | 1.33 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr15_+_52043813 | 1.32 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chrX_+_84499081 | 1.32 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr17_+_64961026 | 1.31 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr22_+_43808014 | 1.30 |
ENST00000334209.5
ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1
|
metallophosphoesterase domain containing 1 |
| chr7_-_142232071 | 1.30 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr19_+_35783028 | 1.29 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chr15_+_52043758 | 1.28 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr12_-_10151773 | 1.27 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr15_+_101402041 | 1.27 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chrX_+_107288280 | 1.26 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_132455118 | 1.26 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr13_-_95131923 | 1.25 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr7_+_139528952 | 1.24 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr5_+_139175380 | 1.24 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr10_-_25241499 | 1.24 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr12_-_11508520 | 1.23 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr5_-_180018540 | 1.23 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr22_+_37959647 | 1.22 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chrX_+_12993336 | 1.20 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr10_+_121578211 | 1.19 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr6_-_127840336 | 1.19 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr2_-_157198860 | 1.19 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_-_143227088 | 1.19 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr17_-_42441204 | 1.18 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr12_-_11463353 | 1.18 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr19_-_51891209 | 1.15 |
ENST00000221973.3
ENST00000596399.1 |
LIM2
|
lens intrinsic membrane protein 2, 19kDa |
| chr14_+_23011142 | 1.15 |
ENST00000390533.1
|
TRAJ4
|
T cell receptor alpha joining 4 |
| chr10_-_99030395 | 1.13 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr20_-_35492048 | 1.13 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr4_-_143226979 | 1.11 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr3_+_133118839 | 1.11 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr20_+_48884002 | 1.11 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr15_+_81293254 | 1.11 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr19_+_35783047 | 1.11 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr12_-_5352315 | 1.11 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr3_-_123339343 | 1.11 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr12_-_49582978 | 1.10 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_-_109777128 | 1.09 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_37899323 | 1.08 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr19_+_35783037 | 1.07 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr6_-_109776901 | 1.07 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr3_-_149510553 | 1.07 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr14_+_100070869 | 1.06 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr6_+_114178512 | 1.04 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr11_+_63974135 | 1.04 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr1_-_6479963 | 1.03 |
ENST00000377836.4
ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2
|
hes family bHLH transcription factor 2 |
| chr20_+_20348740 | 1.03 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr6_+_97372596 | 1.03 |
ENST00000369261.4
|
KLHL32
|
kelch-like family member 32 |
| chr5_-_39424961 | 1.02 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_87028478 | 1.01 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_+_93096619 | 1.00 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr11_+_128563948 | 0.99 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_+_46402583 | 0.99 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_+_1361494 | 0.99 |
ENST00000378821.3
|
TMEM88B
|
transmembrane protein 88B |
| chr5_+_140213815 | 0.99 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr1_-_201476274 | 0.97 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr6_-_64029879 | 0.97 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chrX_+_65384052 | 0.97 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr1_-_201476220 | 0.96 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr12_-_49581152 | 0.95 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr14_+_103589789 | 0.95 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr9_+_139221880 | 0.95 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr12_-_99288536 | 0.95 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_153599578 | 0.94 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr3_-_69402828 | 0.93 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr17_-_9940058 | 0.92 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chr16_+_89989687 | 0.92 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr2_-_114647327 | 0.91 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr17_+_75283973 | 0.91 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr3_-_123339418 | 0.90 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr4_+_2819883 | 0.90 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_+_25205666 | 0.89 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr3_-_181160240 | 0.87 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr13_-_41593425 | 0.87 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr17_-_9939935 | 0.87 |
ENST00000580043.1
|
GAS7
|
growth arrest-specific 7 |
| chr12_+_15475462 | 0.86 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_228337553 | 0.86 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr1_+_212797789 | 0.86 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr11_+_196738 | 0.85 |
ENST00000325113.4
ENST00000342593.5 |
ODF3
|
outer dense fiber of sperm tails 3 |
| chr2_-_70475586 | 0.85 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr16_-_22385901 | 0.84 |
ENST00000268383.2
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa |
| chr2_-_85641162 | 0.84 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr14_-_70038032 | 0.84 |
ENST00000543986.1
|
CCDC177
|
coiled-coil domain containing 177 |
| chr5_+_55033845 | 0.83 |
ENST00000353507.5
ENST00000514278.2 ENST00000505374.1 ENST00000506511.1 |
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr5_-_39425068 | 0.82 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_74610894 | 0.82 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr6_-_49712123 | 0.82 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr1_+_150229554 | 0.81 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr7_+_99006232 | 0.81 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr15_-_80695917 | 0.81 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr17_-_76713100 | 0.80 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr7_-_71868354 | 0.80 |
ENST00000412588.1
|
CALN1
|
calneuron 1 |
| chr19_-_10047219 | 0.80 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr12_+_93096759 | 0.80 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr15_-_73076030 | 0.80 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr5_+_79615790 | 0.79 |
ENST00000296739.4
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chrX_+_65384182 | 0.79 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr12_-_49582593 | 0.78 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_-_103266355 | 0.78 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr21_-_40720995 | 0.77 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chrX_+_107288239 | 0.77 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_125365435 | 0.77 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr22_-_38480100 | 0.76 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr14_+_22982921 | 0.76 |
ENST00000390508.1
|
TRAJ29
|
T cell receptor alpha joining 29 |
| chr10_-_98118724 | 0.75 |
ENST00000393870.2
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr12_-_48213568 | 0.75 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr4_-_103266219 | 0.75 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr4_+_26322987 | 0.74 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_+_3704822 | 0.74 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr12_+_25205628 | 0.74 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr7_-_107968999 | 0.73 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_+_125365110 | 0.73 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr1_+_14075865 | 0.71 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr8_+_27631903 | 0.71 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_+_12859080 | 0.70 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr8_-_20161466 | 0.70 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr2_-_70475701 | 0.70 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.5 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.8 | 14.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.7 | 2.0 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.6 | 1.9 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.6 | 8.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.5 | 2.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.5 | 1.4 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.4 | 2.9 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.4 | 1.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.4 | 3.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.4 | 2.3 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 1.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.4 | 10.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 1.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.3 | 6.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 0.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 4.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 0.9 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 3.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 1.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.2 | 2.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 2.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.6 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.2 | 1.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 2.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 1.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 15.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 2.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 1.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 2.6 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.8 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 2.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.1 | 1.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.2 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 3.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.3 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.7 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.2 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.7 | GO:1905066 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 5.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 2.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.1 | 0.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 1.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 6.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 2.9 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.2 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.8 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 3.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 1.7 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 1.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0035407 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 4.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.6 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 1.1 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.7 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 1.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 2.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.5 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.4 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 3.9 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.7 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.6 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.8 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.7 | 9.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.5 | 2.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 5.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 15.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 2.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 1.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.3 | 1.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 0.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 6.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 0.6 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.2 | 9.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.8 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 1.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 3.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 4.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 9.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.5 | 9.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 1.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 2.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.4 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 3.8 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 1.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.3 | 8.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 1.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 5.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 16.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.2 | 3.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 1.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 1.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 5.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 1.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 3.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.9 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 2.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 12.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 10.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 2.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 2.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 2.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 4.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 8.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 9.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 9.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 4.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 3.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 4.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |