Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
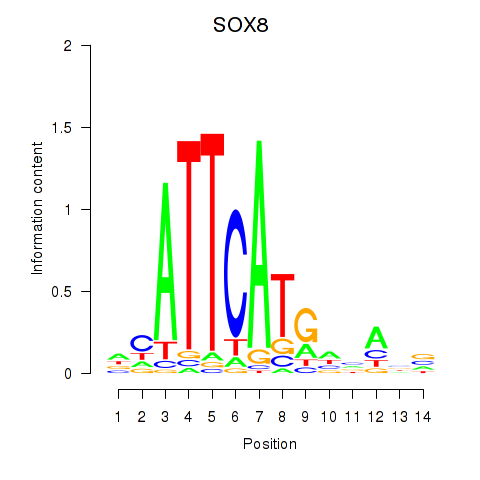
Results for SOX8
Z-value: 1.86
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | 0.64 | 9.1e-05 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_50523843 | 4.41 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr2_-_175711133 | 3.56 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr22_-_50523688 | 3.45 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr5_+_156887027 | 3.12 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr8_-_82359662 | 3.01 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr2_+_26624775 | 2.96 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr4_-_176828307 | 2.74 |
ENST00000513365.1
ENST00000513667.1 ENST00000503563.1 |
GPM6A
|
glycoprotein M6A |
| chr2_+_173792893 | 2.72 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr22_-_50523760 | 2.50 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr16_+_87636474 | 2.47 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr14_+_22337014 | 2.44 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr22_-_50523807 | 2.33 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr5_+_175288631 | 2.31 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_200992827 | 2.21 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr2_+_13677795 | 2.05 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr5_-_160279207 | 2.02 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr7_-_38289173 | 2.01 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr18_-_3845321 | 1.99 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chrX_+_18725758 | 1.96 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr1_+_66820058 | 1.94 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_-_193272741 | 1.92 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr14_+_58797974 | 1.87 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_86253888 | 1.83 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr12_-_10282742 | 1.75 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr20_+_58203664 | 1.73 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_+_36450169 | 1.69 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr9_-_113761720 | 1.69 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr1_+_153330322 | 1.63 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr5_-_160973649 | 1.61 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr6_+_80129989 | 1.60 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr18_-_3845293 | 1.58 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_70944855 | 1.57 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr18_-_5396271 | 1.55 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_167191814 | 1.54 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr12_-_11548496 | 1.53 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr3_-_193272588 | 1.52 |
ENST00000295548.3
|
ATP13A4
|
ATPase type 13A4 |
| chr15_-_89755034 | 1.48 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr12_-_120241187 | 1.48 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chrX_+_100474906 | 1.46 |
ENST00000541709.1
|
DRP2
|
dystrophin related protein 2 |
| chr12_-_10282681 | 1.42 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_+_7118755 | 1.42 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr16_-_30122717 | 1.41 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr12_-_10282836 | 1.41 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr18_-_29264467 | 1.39 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr5_-_94417186 | 1.38 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_-_21699380 | 1.34 |
ENST00000382148.3
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr14_+_52327350 | 1.34 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_+_79371565 | 1.33 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr18_+_56892724 | 1.33 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr1_+_87012753 | 1.33 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr19_-_46999755 | 1.31 |
ENST00000599531.1
|
PNMAL2
|
paraneoplastic Ma antigen family-like 2 |
| chr13_-_47012325 | 1.31 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr16_+_14280564 | 1.30 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_+_38512799 | 1.29 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr5_-_94417314 | 1.29 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_204801471 | 1.27 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr14_+_22580233 | 1.27 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr14_-_106471723 | 1.27 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr3_+_35683816 | 1.26 |
ENST00000438577.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_+_29376673 | 1.25 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr21_+_22519416 | 1.25 |
ENST00000535285.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr7_-_14942283 | 1.25 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr1_+_158901329 | 1.24 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr5_-_94417339 | 1.22 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_120605194 | 1.22 |
ENST00000522167.1
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr10_-_49482907 | 1.19 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr15_+_81299370 | 1.17 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr4_+_159131596 | 1.16 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr1_-_111148241 | 1.15 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr5_+_140186647 | 1.14 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr3_-_39322728 | 1.13 |
ENST00000541347.1
ENST00000412814.1 |
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr4_-_144826682 | 1.13 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr10_-_1779663 | 1.11 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr5_+_140227048 | 1.10 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr14_+_23001452 | 1.10 |
ENST00000390526.1
|
TRAJ11
|
T cell receptor alpha joining 11 |
| chr4_-_144940477 | 1.10 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr16_-_65106110 | 1.09 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr8_+_38244638 | 1.08 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_-_36779411 | 1.07 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr11_+_128563652 | 1.05 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_+_43985084 | 1.05 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr2_-_214017151 | 1.05 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr2_-_158485387 | 1.03 |
ENST00000243349.8
|
ACVR1C
|
activin A receptor, type IC |
| chr11_-_125366089 | 1.03 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chrX_-_75005054 | 1.02 |
ENST00000373359.2
|
MAGEE2
|
melanoma antigen family E, 2 |
| chr12_+_29376592 | 1.02 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr12_+_20848377 | 1.02 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr14_+_75745477 | 1.02 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr16_+_31271274 | 1.01 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr17_-_8021710 | 1.00 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr6_-_32160622 | 1.00 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr2_+_7073174 | 0.99 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr15_+_43885252 | 0.98 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr12_-_11508520 | 0.96 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr11_-_41481135 | 0.96 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr6_+_12718497 | 0.96 |
ENST00000379348.2
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr14_-_106642049 | 0.95 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr1_+_8378140 | 0.94 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr11_-_83878041 | 0.93 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_+_161475208 | 0.93 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr1_-_183538319 | 0.92 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr10_-_87551311 | 0.92 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr4_-_38858428 | 0.92 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr18_-_29264669 | 0.91 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr4_+_156680143 | 0.90 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr15_+_77861183 | 0.90 |
ENST00000560590.1
ENST00000561123.1 |
RP11-307C19.2
|
RP11-307C19.2 |
| chr17_-_16918271 | 0.90 |
ENST00000562897.1
|
RP11-416I2.1
|
RP11-416I2.1 |
| chr6_+_73076432 | 0.90 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_126107042 | 0.89 |
ENST00000535886.1
|
TMEM132B
|
transmembrane protein 132B |
| chr8_-_11873043 | 0.89 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr12_+_56325812 | 0.87 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr22_-_30642728 | 0.86 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr3_+_147657764 | 0.86 |
ENST00000467198.1
ENST00000485006.1 |
RP11-71N10.1
|
RP11-71N10.1 |
| chr3_+_35683651 | 0.86 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr18_-_53253323 | 0.84 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr12_-_95010147 | 0.84 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr3_+_113465866 | 0.84 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr3_+_157154578 | 0.84 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr4_+_40192656 | 0.84 |
ENST00000505618.1
|
RHOH
|
ras homolog family member H |
| chr11_-_112131312 | 0.84 |
ENST00000527122.1
|
C11orf34
|
placenta expressed transcript 1 |
| chr15_+_34261089 | 0.84 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr14_-_106967788 | 0.84 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr12_+_20848282 | 0.83 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_-_50101003 | 0.83 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr12_-_123201337 | 0.83 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr4_-_145061788 | 0.82 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr3_+_35683775 | 0.81 |
ENST00000452563.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_137906154 | 0.80 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr18_-_5577640 | 0.79 |
ENST00000582592.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr4_+_156680153 | 0.78 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr3_-_21792838 | 0.78 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr6_-_133079022 | 0.77 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr22_-_20255212 | 0.77 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr1_-_116383738 | 0.77 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr8_+_35649365 | 0.76 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr5_-_132948216 | 0.75 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr16_+_14280742 | 0.75 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr6_+_148593425 | 0.75 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_-_120354079 | 0.75 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr17_-_34207295 | 0.74 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr10_-_128110441 | 0.74 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr7_-_36634181 | 0.74 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_-_9811600 | 0.74 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr4_-_88450511 | 0.74 |
ENST00000458304.2
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr12_-_50101165 | 0.74 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chr20_-_1472029 | 0.73 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr15_+_77308152 | 0.73 |
ENST00000559859.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr7_+_142012967 | 0.72 |
ENST00000390357.3
|
TRBV4-1
|
T cell receptor beta variable 4-1 |
| chr12_-_39837192 | 0.71 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr6_+_37012607 | 0.71 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr12_+_65277536 | 0.71 |
ENST00000540024.1
|
RP11-766N7.3
|
RP11-766N7.3 |
| chr8_-_126963487 | 0.70 |
ENST00000518964.1
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr17_-_19648683 | 0.70 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_-_153029980 | 0.70 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr14_-_101053739 | 0.69 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr18_-_53253112 | 0.69 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr4_-_100356551 | 0.69 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr7_+_142919130 | 0.69 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr2_-_211342292 | 0.68 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr18_+_61575200 | 0.68 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr12_-_76879852 | 0.67 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr2_+_102413726 | 0.67 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_-_30899924 | 0.66 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr1_-_38512450 | 0.66 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr22_-_50708781 | 0.66 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr12_-_123187890 | 0.66 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr1_+_111415757 | 0.65 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr12_-_57443886 | 0.65 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr2_-_163008903 | 0.65 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr7_-_80551671 | 0.64 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr18_-_53253000 | 0.64 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr4_-_69434245 | 0.64 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chrX_+_15767971 | 0.64 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr15_+_45879964 | 0.64 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr14_+_88471468 | 0.63 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr7_-_64467031 | 0.63 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr11_-_128457446 | 0.63 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr6_-_116833500 | 0.62 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr6_-_136788001 | 0.62 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr7_+_139478030 | 0.61 |
ENST00000425687.1
ENST00000263552.6 ENST00000438104.1 ENST00000336425.5 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr16_+_57769635 | 0.61 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr19_+_18794470 | 0.60 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr2_-_187367356 | 0.60 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr19_+_1269324 | 0.60 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr11_-_129062093 | 0.59 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr15_+_78830023 | 0.59 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr20_+_42543504 | 0.59 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr6_+_147527103 | 0.58 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chrX_-_50386648 | 0.58 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr6_-_10115007 | 0.58 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr6_+_31021225 | 0.58 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr8_+_30496078 | 0.56 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr8_+_21915368 | 0.56 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr12_-_10007448 | 0.56 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr20_-_1472053 | 0.55 |
ENST00000537284.1
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr8_-_126963417 | 0.54 |
ENST00000500989.2
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr14_+_23000889 | 0.53 |
ENST00000390525.1
|
TRAJ12
|
T cell receptor alpha joining 12 |
| chr10_-_38265517 | 0.53 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr2_-_239140011 | 0.52 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr6_-_138539627 | 0.51 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.6 | 1.7 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.5 | 1.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 1.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.3 | 1.0 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.3 | 1.7 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 1.9 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 0.9 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.3 | 2.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.3 | 0.8 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.3 | 1.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 0.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 1.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.7 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 4.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.2 | 0.6 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 0.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 3.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 0.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 2.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.5 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.7 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.4 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.3 | GO:0006174 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 3.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.3 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 2.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.3 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.7 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 1.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 3.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 1.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.1 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 0.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.1 | 0.9 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 2.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 2.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 1.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 1.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 1.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 2.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 2.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 1.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 1.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.4 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.4 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.5 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.8 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0052361 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.9 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 1.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.4 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.6 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 2.0 | GO:0048636 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) |
| 0.0 | 0.2 | GO:1903416 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) response to glycoside(GO:1903416) |
| 0.0 | 2.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 3.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 2.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 3.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 1.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 2.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.2 | 0.9 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 1.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 1.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.6 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.1 | 1.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 4.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 3.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 11.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 2.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 2.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 7.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 1.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.3 | 1.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 0.9 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.3 | 0.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 0.8 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 1.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 1.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.0 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 0.7 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 2.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 1.7 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.6 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 1.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 3.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 2.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 3.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 3.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.1 | 2.5 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 13.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 3.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 3.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 2.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 3.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.0 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 2.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 2.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.2 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.7 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 3.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 4.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 5.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |