Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
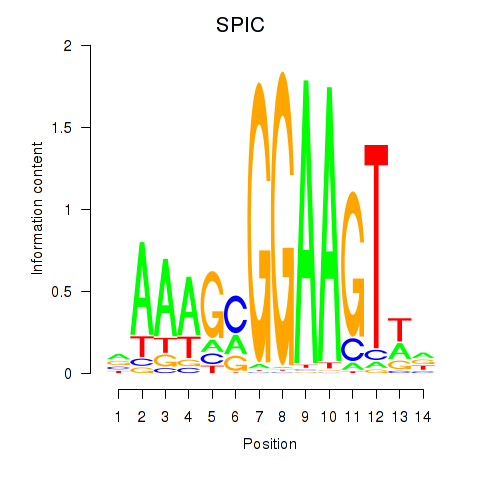
Results for SPIC
Z-value: 4.54
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIC | hg19_v2_chr12_+_101869096_101869199 | 0.15 | 4.0e-01 | Click! |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_72709050 | 12.99 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr12_+_54892550 | 11.24 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr4_-_74853897 | 11.21 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr19_+_42381337 | 10.71 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr1_+_159772121 | 10.46 |
ENST00000339348.5
ENST00000392235.3 ENST00000368106.3 |
FCRL6
|
Fc receptor-like 6 |
| chr19_-_8642289 | 10.46 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
| chr6_+_31554826 | 10.00 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr20_+_48892848 | 9.89 |
ENST00000422459.1
|
RP11-290F20.3
|
RP11-290F20.3 |
| chr1_-_157789850 | 9.59 |
ENST00000491942.1
ENST00000358292.3 ENST00000368176.3 |
FCRL1
|
Fc receptor-like 1 |
| chr14_+_88471468 | 9.48 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr1_-_160681593 | 9.06 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr1_+_161677034 | 9.02 |
ENST00000349527.4
ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA
|
Fc receptor-like A |
| chrX_+_78200829 | 8.98 |
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chrX_+_78200913 | 8.90 |
ENST00000171757.2
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr1_+_192127578 | 8.86 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr19_-_10446449 | 8.56 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_+_30194916 | 8.39 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr7_-_142099977 | 8.30 |
ENST00000390359.3
|
TRBV7-8
|
T cell receptor beta variable 7-8 |
| chr6_-_159466136 | 8.29 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr16_+_30485267 | 8.24 |
ENST00000569725.1
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chrX_-_47489244 | 8.21 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr1_+_161676983 | 8.20 |
ENST00000367957.2
|
FCRLA
|
Fc receptor-like A |
| chr22_+_37309662 | 8.11 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr14_-_23284703 | 7.97 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_-_9885888 | 7.97 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr11_-_58980342 | 7.68 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr6_-_133079022 | 7.66 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr6_-_159466042 | 7.34 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr1_-_25256368 | 7.34 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr1_-_157746909 | 7.34 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr14_-_23284675 | 7.31 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr15_-_90358048 | 7.29 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr19_-_54784937 | 7.20 |
ENST00000434421.1
ENST00000314446.5 ENST00000391749.4 |
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr19_-_39108568 | 7.19 |
ENST00000586296.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr17_-_72619869 | 7.16 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr6_+_31554612 | 7.10 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr17_-_72619783 | 7.03 |
ENST00000328630.3
|
CD300E
|
CD300e molecule |
| chr17_+_72462525 | 7.00 |
ENST00000360141.3
|
CD300A
|
CD300a molecule |
| chr5_-_66492562 | 6.94 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr19_-_51875894 | 6.85 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr6_+_106534192 | 6.82 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr19_+_49838653 | 6.75 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr17_-_72527605 | 6.72 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr2_+_182321925 | 6.67 |
ENST00000339307.4
ENST00000397033.2 |
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr19_+_55084438 | 6.66 |
ENST00000439534.1
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
| chr11_-_118083600 | 6.52 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr9_+_214842 | 6.50 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr20_-_1638408 | 6.49 |
ENST00000303415.3
ENST00000381583.2 |
SIRPG
|
signal-regulatory protein gamma |
| chr1_+_161676739 | 6.42 |
ENST00000236938.6
ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA
|
Fc receptor-like A |
| chr2_+_89998789 | 6.40 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr9_+_215158 | 6.38 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chrX_-_70838306 | 6.37 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chr6_+_31554779 | 6.36 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr19_-_39108643 | 6.33 |
ENST00000396857.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr8_+_11351494 | 6.31 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr7_+_150382781 | 6.27 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr22_+_23247030 | 6.20 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr12_-_9885702 | 6.14 |
ENST00000542530.1
|
CLECL1
|
C-type lectin-like 1 |
| chr10_-_98480243 | 6.13 |
ENST00000339364.5
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr13_-_46756351 | 6.11 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_+_89999259 | 6.11 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr20_-_1638360 | 6.09 |
ENST00000216927.4
ENST00000344103.4 |
SIRPG
|
signal-regulatory protein gamma |
| chr14_-_23285011 | 6.01 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr19_-_39108552 | 5.97 |
ENST00000591517.1
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr7_+_74188309 | 5.96 |
ENST00000289473.4
ENST00000433458.1 |
NCF1
|
neutrophil cytosolic factor 1 |
| chr6_-_41122063 | 5.89 |
ENST00000426005.2
ENST00000437044.2 ENST00000373127.4 |
TREML1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr1_+_111415757 | 5.89 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr6_+_391739 | 5.88 |
ENST00000380956.4
|
IRF4
|
interferon regulatory factor 4 |
| chr19_+_51645556 | 5.86 |
ENST00000601682.1
ENST00000317643.6 ENST00000305628.7 ENST00000600577.1 |
SIGLEC7
|
sialic acid binding Ig-like lectin 7 |
| chr19_-_52307357 | 5.81 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr3_+_32433363 | 5.74 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_-_150738261 | 5.74 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr6_+_31554636 | 5.73 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr17_+_72462766 | 5.70 |
ENST00000392625.3
ENST00000361933.3 ENST00000310828.5 |
CD300A
|
CD300a molecule |
| chr22_+_37257015 | 5.64 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr17_-_37934466 | 5.52 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr12_+_54891495 | 5.50 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr14_-_23285069 | 5.40 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_+_31554456 | 5.34 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr19_-_10450287 | 5.31 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr3_+_32433154 | 5.25 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr7_-_115799942 | 5.25 |
ENST00000484212.1
|
TFEC
|
transcription factor EC |
| chr1_-_160549235 | 5.22 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr12_+_7055767 | 5.21 |
ENST00000447931.2
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_-_149465990 | 5.21 |
ENST00000543093.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr16_+_81812863 | 5.20 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr1_-_161519579 | 5.17 |
ENST00000426740.1
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr9_+_273038 | 5.17 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr19_-_7766991 | 5.17 |
ENST00000597921.1
ENST00000346664.5 |
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr16_+_57576584 | 5.15 |
ENST00000340339.4
|
GPR114
|
G protein-coupled receptor 114 |
| chr22_-_37880543 | 5.15 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_183560011 | 5.09 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr17_-_29641104 | 5.06 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_40810516 | 5.05 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr17_+_34431212 | 5.05 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr2_+_218994002 | 5.04 |
ENST00000428565.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr14_+_21423611 | 5.03 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr17_-_29641084 | 5.03 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr12_-_8218926 | 5.01 |
ENST00000546241.1
|
C3AR1
|
complement component 3a receptor 1 |
| chr12_+_25205666 | 4.99 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr19_-_10450328 | 4.97 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_+_233924548 | 4.95 |
ENST00000422935.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_+_35939154 | 4.95 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr16_-_21663919 | 4.93 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr1_-_161519682 | 4.93 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr3_+_52321827 | 4.90 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr11_-_59951430 | 4.85 |
ENST00000533409.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr4_-_103682071 | 4.83 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr10_+_26727333 | 4.80 |
ENST00000356785.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr16_-_21663950 | 4.79 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr16_+_31271274 | 4.74 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr2_-_64568781 | 4.69 |
ENST00000424119.1
|
AC114752.3
|
AC114752.3 |
| chr12_-_8218997 | 4.60 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr1_+_241695424 | 4.58 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr21_-_15918618 | 4.57 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr12_+_7055631 | 4.54 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_-_20370847 | 4.52 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr5_+_96211643 | 4.52 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr12_-_15114603 | 4.50 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr20_+_48884002 | 4.50 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr1_+_161475208 | 4.49 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr12_+_105724613 | 4.47 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr2_-_89545079 | 4.45 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr19_-_51920835 | 4.43 |
ENST00000442846.3
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_-_15114658 | 4.42 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_+_6772710 | 4.41 |
ENST00000304076.2
ENST00000602142.1 ENST00000596764.1 |
VAV1
|
vav 1 guanine nucleotide exchange factor |
| chr6_+_36973406 | 4.39 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr2_+_113885138 | 4.35 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr12_+_25205628 | 4.34 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_8608522 | 4.28 |
ENST00000382073.3
|
CLEC6A
|
C-type lectin domain family 6, member A |
| chr1_+_196912902 | 4.28 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr15_-_80263506 | 4.26 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr4_+_68424434 | 4.25 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr6_+_31553978 | 4.24 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr5_+_57787254 | 4.23 |
ENST00000502276.1
ENST00000396776.2 ENST00000511930.1 |
GAPT
|
GRB2-binding adaptor protein, transmembrane |
| chr2_+_202125219 | 4.21 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr12_+_47610315 | 4.20 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr3_+_121796697 | 4.19 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr19_-_54804173 | 4.18 |
ENST00000391744.3
ENST00000251390.3 |
LILRA3
|
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr1_-_31230650 | 4.17 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr16_-_68034452 | 4.17 |
ENST00000575510.1
|
DPEP2
|
dipeptidase 2 |
| chr15_+_77287426 | 4.16 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr19_-_6481759 | 4.15 |
ENST00000588421.1
|
DENND1C
|
DENN/MADD domain containing 1C |
| chr19_+_35940486 | 4.13 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr7_+_150264365 | 4.13 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr20_-_1569278 | 4.10 |
ENST00000262929.5
ENST00000567028.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr6_-_6225026 | 4.08 |
ENST00000445223.1
|
F13A1
|
coagulation factor XIII, A1 polypeptide |
| chr1_+_161551101 | 4.06 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr3_-_121379739 | 4.05 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_+_31309645 | 4.04 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr19_-_36233332 | 4.04 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr4_+_8201091 | 3.99 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_+_161632937 | 3.98 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr5_-_180235755 | 3.97 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr6_+_31582961 | 3.94 |
ENST00000376059.3
ENST00000337917.7 |
AIF1
|
allograft inflammatory factor 1 |
| chr17_-_72542278 | 3.93 |
ENST00000330793.1
|
CD300C
|
CD300c molecule |
| chr4_+_74606223 | 3.90 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr19_-_6481776 | 3.88 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr18_-_61089611 | 3.84 |
ENST00000591519.1
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr1_+_149230680 | 3.83 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr19_-_14887568 | 3.82 |
ENST00000596991.2
ENST00000594294.1 ENST00000594076.1 ENST00000595839.1 ENST00000392965.3 ENST00000601345.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr21_-_46340807 | 3.80 |
ENST00000397846.3
ENST00000524251.1 ENST00000522688.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_143886877 | 3.76 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr17_+_7482785 | 3.76 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr19_-_18508396 | 3.74 |
ENST00000595840.1
ENST00000339007.3 |
LRRC25
|
leucine rich repeat containing 25 |
| chr21_-_46340884 | 3.74 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_-_51920873 | 3.71 |
ENST00000441969.3
ENST00000525998.1 ENST00000436984.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr19_-_39826639 | 3.71 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr2_+_181988620 | 3.70 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr19_+_46010674 | 3.70 |
ENST00000245932.6
ENST00000592139.1 ENST00000590603.1 |
VASP
|
vasodilator-stimulated phosphoprotein |
| chr11_+_64107663 | 3.69 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr16_-_88717482 | 3.69 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr19_+_55128576 | 3.68 |
ENST00000396331.1
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr13_+_43148281 | 3.68 |
ENST00000239849.6
ENST00000398795.2 ENST00000544862.1 |
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11 |
| chr8_+_27169138 | 3.68 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr15_+_77287715 | 3.67 |
ENST00000559161.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr14_-_106114739 | 3.66 |
ENST00000460164.1
|
RP11-731F5.2
|
RP11-731F5.2 |
| chr19_-_51920952 | 3.65 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_+_7060508 | 3.65 |
ENST00000541698.1
ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrY_+_2709527 | 3.64 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr1_+_112016414 | 3.62 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_-_157522180 | 3.61 |
ENST00000356953.4
ENST00000368188.2 ENST00000368190.3 ENST00000368189.3 |
FCRL5
|
Fc receptor-like 5 |
| chr16_+_66613351 | 3.59 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr1_-_157522260 | 3.58 |
ENST00000368191.3
ENST00000361835.3 |
FCRL5
|
Fc receptor-like 5 |
| chr14_-_106518922 | 3.58 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr1_+_117544366 | 3.55 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr1_-_183559693 | 3.55 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_-_113594279 | 3.53 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr20_+_43595115 | 3.53 |
ENST00000372806.3
ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4
|
serine/threonine kinase 4 |
| chr16_-_88851618 | 3.46 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr5_-_140013224 | 3.45 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr3_+_52813932 | 3.45 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr20_-_3687775 | 3.43 |
ENST00000344754.4
ENST00000202578.4 |
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr19_-_52133588 | 3.42 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr5_-_78281603 | 3.38 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr12_+_7060414 | 3.37 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_-_164534657 | 3.36 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr19_+_55141861 | 3.36 |
ENST00000396327.3
ENST00000324602.7 ENST00000434867.2 |
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr5_-_140013275 | 3.36 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr19_+_35645618 | 3.34 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.7 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 6.0 | 18.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 4.5 | 13.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 4.2 | 16.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 3.4 | 10.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 3.3 | 13.2 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 3.2 | 18.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 3.1 | 31.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 3.0 | 9.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 2.5 | 10.1 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 2.3 | 7.0 | GO:0061582 | colon epithelial cell migration(GO:0061580) intestinal epithelial cell migration(GO:0061582) |
| 2.3 | 11.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.3 | 13.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 2.3 | 6.8 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 2.2 | 6.7 | GO:0050904 | diapedesis(GO:0050904) |
| 2.2 | 11.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 2.1 | 14.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 2.0 | 10.1 | GO:0039513 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 1.9 | 5.8 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.8 | 7.2 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 1.8 | 1.8 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 1.7 | 8.4 | GO:0032796 | uropod organization(GO:0032796) |
| 1.6 | 6.3 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 1.4 | 10.0 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.4 | 4.3 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 1.4 | 7.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.3 | 5.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 1.3 | 3.8 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 1.2 | 3.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.2 | 3.7 | GO:0071810 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 1.2 | 3.5 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 1.2 | 3.5 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.2 | 3.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.1 | 3.2 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 1.0 | 7.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.0 | 5.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 1.0 | 3.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 1.0 | 13.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.9 | 9.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 5.7 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.9 | 22.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.9 | 6.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.9 | 2.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.9 | 15.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.9 | 6.8 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.8 | 5.0 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.8 | 4.9 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.8 | 7.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 22.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.8 | 2.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.8 | 5.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.7 | 5.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.7 | 3.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.7 | 4.3 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.7 | 2.8 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.7 | 13.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 1.3 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.7 | 4.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.6 | 0.6 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.6 | 23.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.6 | 3.7 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.6 | 1.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.6 | 3.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.6 | 1.7 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.6 | 2.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.6 | 3.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.6 | 3.9 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.6 | 1.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.6 | 19.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 9.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.5 | 2.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.5 | 5.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.5 | 2.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.5 | 2.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.5 | 1.5 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.5 | 6.0 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.5 | 1.5 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.5 | 1.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.5 | 2.9 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.5 | 1.4 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.5 | 1.4 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.4 | 3.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.4 | 4.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.4 | 2.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.4 | 0.9 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.4 | 1.7 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.4 | 1.7 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.4 | 5.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.4 | 2.5 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.4 | 2.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 1.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 0.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.4 | 1.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.4 | 7.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.4 | 13.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.4 | 1.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.4 | 2.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 2.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.4 | 4.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 2.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 3.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 2.9 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.3 | 1.0 | GO:0072717 | response to actinomycin D(GO:0072716) cellular response to actinomycin D(GO:0072717) |
| 0.3 | 3.8 | GO:1902961 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.3 | 2.4 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 1.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 3.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.3 | 3.4 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.3 | 2.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.3 | 5.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 1.9 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 33.6 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.3 | 1.6 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 4.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 5.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 3.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.4 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.3 | 6.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 6.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.3 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.3 | 1.9 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.3 | 0.8 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.3 | 5.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 1.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 2.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 3.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.2 | 2.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 3.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 1.9 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 4.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 0.7 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.2 | 1.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.3 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 50.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 0.6 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.2 | 5.8 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.2 | 3.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 4.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 6.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 1.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.8 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.8 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.2 | 2.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.8 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.2 | 2.0 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 2.8 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.2 | 1.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.2 | 4.7 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 | 1.0 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 3.5 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 | 2.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 1.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 1.3 | GO:0090023 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0010628 | positive regulation of gene expression(GO:0010628) |
| 0.2 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 13.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 0.7 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) notochord regression(GO:0060032) |
| 0.2 | 1.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 6.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.2 | 1.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 3.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.2 | 5.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 1.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 2.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 0.5 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.2 | 0.8 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 1.4 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.4 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 13.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 0.6 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 1.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.8 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.0 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 8.8 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 0.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 1.0 | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002923) |
| 0.1 | 1.3 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 3.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.4 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 4.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 8.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 5.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.1 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 0.5 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.1 | 2.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 2.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 3.8 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.1 | 0.9 | GO:0051270 | regulation of cellular component movement(GO:0051270) |
| 0.1 | 1.8 | GO:0070857 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) regulation of bile acid biosynthetic process(GO:0070857) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 19.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.9 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 1.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 2.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 1.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.9 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.5 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.1 | 4.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 1.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 1.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.6 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 1.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 16.5 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.1 | 0.7 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 6.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.1 | 2.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 2.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 5.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.7 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.1 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 5.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.6 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 1.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 2.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 7.2 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.1 | 0.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 9.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 3.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 5.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.9 | GO:1902593 | single-organism nuclear import(GO:1902593) |
| 0.1 | 2.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.5 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 1.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 1.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.7 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.1 | 1.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.3 | GO:1902513 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.7 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 7.6 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 2.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 12.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 15.8 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.9 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 1.8 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 2.7 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 1.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.7 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.5 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.4 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.2 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 4.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 25.1 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 3.8 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 6.5 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 2.7 | GO:0050870 | positive regulation of T cell activation(GO:0050870) |
| 0.0 | 1.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 3.2 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.7 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 2.4 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.3 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 3.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 3.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 2.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.7 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 1.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 1.3 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.9 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 1.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0090283 | negative regulation of protein glycosylation(GO:0060051) regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.3 | GO:1903203 | regulation of oxidative stress-induced neuron death(GO:1903203) negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.1 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.8 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.4 | GO:0045595 | regulation of cell differentiation(GO:0045595) |
| 0.0 | 2.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.6 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.1 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 2.1 | 8.2 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 2.1 | 20.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.9 | 15.5 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 1.5 | 20.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.5 | 10.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.5 | 13.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.4 | 5.7 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 1.3 | 6.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 1.1 | 20.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 5.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.7 | 5.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 2.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.7 | 2.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.6 | 2.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.6 | 2.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.5 | 3.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.5 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 10.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 3.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.5 | 11.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.5 | 16.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 1.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 7.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 3.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 10.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 2.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 1.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.4 | 43.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.4 | 1.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.4 | 0.8 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.4 | 1.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.4 | 5.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 31.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.3 | 1.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.3 | 1.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 3.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 1.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 0.9 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.3 | 6.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 1.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 12.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 4.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.2 | 1.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 4.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 0.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 2.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 2.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.8 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.2 | 76.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 17.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 1.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 0.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 7.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 4.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 2.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 2.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.9 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 2.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 3.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 17.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 4.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 2.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 12.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 3.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 7.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 9.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 1.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 7.9 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 13.5 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 6.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 3.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 17.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 11.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.4 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 14.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 10.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 12.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 11.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 7.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 7.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 107.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 38.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 3.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.4 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 2.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 3.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 15.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 3.8 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.9 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 1.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.8 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 8.0 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0030684 | preribosome(GO:0030684) small-subunit processome(GO:0032040) |
| 0.0 | 3.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 88.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0044454 | nuclear chromosome part(GO:0044454) |
| 0.0 | 1.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.6 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 2.5 | 10.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 2.2 | 11.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 1.8 | 7.2 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 1.8 | 12.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.8 | 7.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 1.6 | 8.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.6 | 19.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 1.6 | 4.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.5 | 4.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.4 | 10.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.4 | 11.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 1.4 | 10.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.3 | 5.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.3 | 29.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.3 | 3.8 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 1.3 | 7.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.2 | 14.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.2 | 3.5 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 1.1 | 3.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.1 | 20.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.0 | 5.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 1.0 | 14.6 | GO:0019864 | IgG binding(GO:0019864) |
| 1.0 | 5.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.0 | 2.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.9 | 15.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.8 | 5.0 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.8 | 3.2 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.8 | 22.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.8 | 4.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.8 | 12.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 19.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 12.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 5.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.7 | 6.8 | GO:0005497 | androgen binding(GO:0005497) |
| 0.7 | 6.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.7 | 2.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.6 | 6.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.6 | 3.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.6 | 2.4 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.6 | 23.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 5.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.6 | 5.7 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.6 | 3.4 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.6 | 2.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.6 | 1.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.6 | 2.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 1.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.5 | 4.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 2.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.5 | 2.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.5 | 4.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.5 | 2.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.5 | 1.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.5 | 10.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 5.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.5 | 6.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 1.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.5 | 2.8 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.5 | 1.4 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.4 | 4.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.4 | 1.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.4 | 1.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.4 | 1.3 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.4 | 8.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.4 | 11.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.4 | 12.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 3.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.4 | 2.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 2.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.4 | 2.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 3.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 5.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 1.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.3 | 1.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 3.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 0.9 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.3 | 4.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 2.4 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 0.9 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 8.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 1.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 6.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 3.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 3.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 2.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 0.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 0.8 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.3 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.0 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.2 | 2.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 1.0 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 2.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.7 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.9 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 4.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 3.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 1.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 0.6 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.2 | 2.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 0.8 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 7.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 4.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 0.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 10.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 22.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 2.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 1.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 3.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 9.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.7 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 1.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 4.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 32.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 2.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.5 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 2.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.0 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 5.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 1.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 6.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 4.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 46.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.6 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 1.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 2.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 4.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 2.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 2.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 5.4 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 3.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 6.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 8.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.9 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 14.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 3.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.9 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 1.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.0 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 1.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 29.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 4.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.7 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.2 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 4.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.3 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 14.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 4.2 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 1.5 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 2.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 4.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 2.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 45.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 92.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.6 | 9.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 16.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 12.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 11.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 4.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 30.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 10.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 3.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 6.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 8.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 11.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 10.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 7.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 9.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 7.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 5.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 8.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 19.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 11.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 18.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 5.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 10.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 5.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 2.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 11.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 6.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 13.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 5.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 21.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.1 | 47.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.9 | 21.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 13.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.7 | 85.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 10.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 46.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.6 | 41.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.5 | 7.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.5 | 29.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 7.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 20.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.4 | 7.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 1.9 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.3 | 5.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 6.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 11.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 4.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 6.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 4.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 16.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 5.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 4.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 3.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.2 | 4.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 2.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 7.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 1.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 2.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 2.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 5.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 21.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 8.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.8 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 34.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.9 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.1 | 3.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 25.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 2.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 7.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 7.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 7.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 4.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 2.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 5.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 5.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.6 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |