Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for STAT5B
Z-value: 2.00
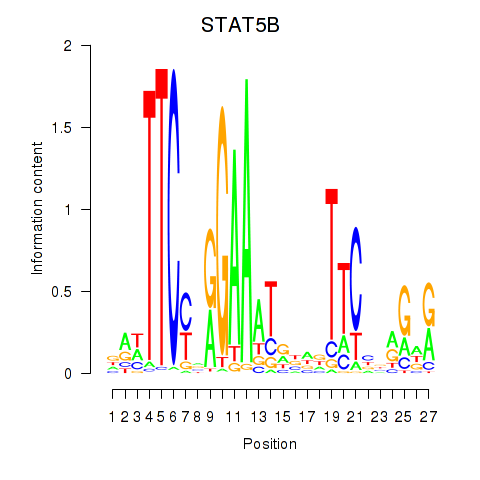
Transcription factors associated with STAT5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5B
|
ENSG00000173757.5 | signal transducer and activator of transcription 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5B | hg19_v2_chr17_-_40428359_40428462 | -0.23 | 2.0e-01 | Click! |
Activity profile of STAT5B motif
Sorted Z-values of STAT5B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_50523843 | 2.85 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_-_151047327 | 2.84 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr22_-_50523807 | 2.78 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50523688 | 2.58 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_-_26697304 | 2.31 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr1_+_149754227 | 2.16 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr22_-_50523760 | 2.09 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_37213554 | 1.98 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr6_+_116782527 | 1.95 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr12_+_21207503 | 1.94 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_-_55367361 | 1.93 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr2_+_17721230 | 1.90 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr12_-_55367331 | 1.88 |
ENST00000526532.1
ENST00000532757.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr4_+_155484155 | 1.85 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr19_+_44556158 | 1.85 |
ENST00000434772.3
ENST00000585552.1 |
ZNF223
|
zinc finger protein 223 |
| chr4_-_155511887 | 1.85 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr4_+_155484103 | 1.83 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr1_-_120935894 | 1.80 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr11_+_27062860 | 1.74 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_-_104145795 | 1.73 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chrX_+_107288280 | 1.67 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_-_44405623 | 1.52 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr5_-_111093081 | 1.51 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr7_+_110731062 | 1.49 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr11_+_27062502 | 1.47 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_6051526 | 1.46 |
ENST00000378111.1
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_-_49016847 | 1.42 |
ENST00000598924.1
|
CTC-273B12.10
|
CTC-273B12.10 |
| chr18_-_67629015 | 1.40 |
ENST00000579496.1
|
CD226
|
CD226 molecule |
| chr3_-_130745403 | 1.39 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr6_-_135818764 | 1.38 |
ENST00000488690.2
|
AHI1
|
Abelson helper integration site 1 |
| chr1_+_159770292 | 1.37 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr19_+_44488330 | 1.36 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr19_+_51152702 | 1.34 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr14_+_22180536 | 1.28 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr5_-_111093167 | 1.28 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr7_-_134001663 | 1.26 |
ENST00000378509.4
|
SLC35B4
|
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
| chr19_+_44617511 | 1.26 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr9_-_99637820 | 1.24 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr5_-_111092930 | 1.22 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr19_+_44669280 | 1.21 |
ENST00000590089.1
ENST00000454662.2 |
ZNF226
|
zinc finger protein 226 |
| chr19_+_44529479 | 1.17 |
ENST00000587846.1
ENST00000187879.8 ENST00000391960.3 |
ZNF222
|
zinc finger protein 222 |
| chr12_+_120740119 | 1.11 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr19_-_58326267 | 1.11 |
ENST00000391701.1
|
ZNF552
|
zinc finger protein 552 |
| chr6_+_52226897 | 1.09 |
ENST00000442253.2
|
PAQR8
|
progestin and adipoQ receptor family member VIII |
| chr3_-_130745571 | 1.06 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr19_+_44507091 | 1.06 |
ENST00000429154.2
ENST00000585632.1 |
ZNF230
|
zinc finger protein 230 |
| chr7_+_47834880 | 1.05 |
ENST00000258776.4
|
C7orf69
|
chromosome 7 open reading frame 69 |
| chr5_-_111092873 | 1.03 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr16_+_69984810 | 1.00 |
ENST00000393701.2
ENST00000568461.1 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr1_+_155051305 | 0.99 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr6_+_28048753 | 0.99 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr19_-_3801789 | 0.97 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chrX_-_70474910 | 0.96 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr5_-_111093340 | 0.95 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr14_-_68162464 | 0.95 |
ENST00000553384.1
ENST00000557726.1 ENST00000381346.4 |
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr19_-_45004556 | 0.95 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr19_+_35225060 | 0.94 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr19_+_45445491 | 0.94 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr12_+_14369524 | 0.93 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr11_-_82997420 | 0.93 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr19_-_44439387 | 0.92 |
ENST00000269973.5
|
ZNF45
|
zinc finger protein 45 |
| chr19_+_38085768 | 0.92 |
ENST00000316433.4
ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540
|
zinc finger protein 540 |
| chr19_-_56904799 | 0.92 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr19_+_58258164 | 0.90 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr3_-_181160240 | 0.90 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr19_+_40503013 | 0.90 |
ENST00000595225.1
|
ZNF546
|
zinc finger protein 546 |
| chr1_-_112106556 | 0.90 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr16_+_70207686 | 0.90 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr19_+_38085731 | 0.89 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr15_+_69854027 | 0.89 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr16_+_76343743 | 0.88 |
ENST00000478060.1
|
CNTNAP4
|
contactin associated protein-like 4 |
| chr9_+_104161123 | 0.88 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chrY_+_2709906 | 0.88 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr9_+_123906331 | 0.86 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr2_-_170430366 | 0.86 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr3_+_156799587 | 0.86 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr1_-_112106578 | 0.85 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr19_+_44669221 | 0.85 |
ENST00000590578.1
ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226
|
zinc finger protein 226 |
| chrX_+_107288197 | 0.84 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_+_10197463 | 0.84 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr14_-_68162438 | 0.82 |
ENST00000557273.1
ENST00000428130.2 |
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr2_-_170430277 | 0.81 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr19_+_40502938 | 0.81 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr12_+_21284118 | 0.81 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr2_-_207583120 | 0.80 |
ENST00000452335.2
|
DYTN
|
dystrotelin |
| chr19_-_58090240 | 0.80 |
ENST00000196489.3
|
ZNF416
|
zinc finger protein 416 |
| chr6_-_25874440 | 0.79 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr1_+_40997233 | 0.79 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr12_+_133656995 | 0.79 |
ENST00000356456.5
|
ZNF140
|
zinc finger protein 140 |
| chr19_-_44405941 | 0.78 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr8_+_93895865 | 0.77 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr19_+_58038683 | 0.77 |
ENST00000240719.3
ENST00000376233.3 ENST00000594943.1 ENST00000602149.1 |
ZNF549
|
zinc finger protein 549 |
| chrY_+_2709527 | 0.76 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr1_+_155051379 | 0.74 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr20_+_34556488 | 0.74 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr4_+_74347400 | 0.74 |
ENST00000226355.3
|
AFM
|
afamin |
| chr3_-_183735651 | 0.74 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chrX_+_107288239 | 0.73 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr19_-_37406923 | 0.73 |
ENST00000520965.1
|
ZNF829
|
zinc finger protein 829 |
| chr13_+_52436111 | 0.71 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr19_-_4902877 | 0.70 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr19_+_35225121 | 0.70 |
ENST00000595708.1
ENST00000593781.1 |
ZNF181
|
zinc finger protein 181 |
| chr12_+_133657461 | 0.67 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr19_+_44598534 | 0.66 |
ENST00000336976.6
|
ZNF224
|
zinc finger protein 224 |
| chr19_+_13229126 | 0.66 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_-_16472483 | 0.66 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr19_+_15783879 | 0.65 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chr19_-_38183201 | 0.65 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr6_-_27440460 | 0.63 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr2_-_37193606 | 0.62 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr9_-_115774453 | 0.61 |
ENST00000427548.1
|
ZNF883
|
zinc finger protein 883 |
| chr1_-_168464875 | 0.60 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_-_37407172 | 0.60 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr19_-_37958318 | 0.59 |
ENST00000316950.6
ENST00000591710.1 |
ZNF569
|
zinc finger protein 569 |
| chr19_-_57988871 | 0.59 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr16_-_20710212 | 0.58 |
ENST00000523065.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr9_+_100069933 | 0.58 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr19_-_37157713 | 0.57 |
ENST00000592829.1
ENST00000591370.1 ENST00000588268.1 ENST00000360357.4 |
ZNF461
|
zinc finger protein 461 |
| chr9_+_130186653 | 0.57 |
ENST00000342483.5
ENST00000543471.1 |
ZNF79
|
zinc finger protein 79 |
| chr6_+_135818979 | 0.56 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr17_+_15603447 | 0.56 |
ENST00000395893.2
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr10_-_127505167 | 0.56 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr2_-_214017151 | 0.55 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_-_146054494 | 0.54 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr11_-_87908600 | 0.54 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr6_+_84569359 | 0.53 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr13_-_99404875 | 0.53 |
ENST00000376503.5
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr11_+_73357614 | 0.53 |
ENST00000536527.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr16_-_53737795 | 0.53 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr19_+_44598492 | 0.52 |
ENST00000589680.1
|
ZNF224
|
zinc finger protein 224 |
| chr2_+_63816295 | 0.52 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr6_-_27440837 | 0.52 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr7_-_93633658 | 0.52 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr14_-_77889860 | 0.52 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chrX_+_106045891 | 0.51 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr20_+_54987305 | 0.51 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr2_+_63816269 | 0.51 |
ENST00000432309.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr9_-_104160872 | 0.51 |
ENST00000539624.1
ENST00000374865.4 |
MRPL50
|
mitochondrial ribosomal protein L50 |
| chr2_+_63816126 | 0.50 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr15_+_22833395 | 0.49 |
ENST00000283645.4
|
TUBGCP5
|
tubulin, gamma complex associated protein 5 |
| chr13_+_115079949 | 0.49 |
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr7_-_93633684 | 0.49 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr2_+_172290707 | 0.49 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr11_+_2920951 | 0.49 |
ENST00000347936.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr2_+_27760247 | 0.49 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr7_+_99102267 | 0.49 |
ENST00000326775.5
ENST00000451158.1 |
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr19_+_58258202 | 0.48 |
ENST00000431353.1
|
ZNF776
|
zinc finger protein 776 |
| chr6_+_2245982 | 0.47 |
ENST00000530346.1
ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr19_-_44905741 | 0.46 |
ENST00000591679.1
ENST00000544719.2 ENST00000330997.4 ENST00000585868.1 ENST00000588212.1 |
ZNF285
CTC-512J12.6
|
zinc finger protein 285 Uncharacterized protein |
| chr11_-_82997371 | 0.46 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr19_+_13228917 | 0.46 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr2_+_63816087 | 0.46 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr8_+_27632047 | 0.44 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr4_+_40195262 | 0.44 |
ENST00000503941.1
|
RHOH
|
ras homolog family member H |
| chr19_-_36980337 | 0.44 |
ENST00000434377.2
ENST00000424129.2 ENST00000452939.1 ENST00000427002.1 |
ZNF566
|
zinc finger protein 566 |
| chr15_+_22833482 | 0.43 |
ENST00000453949.2
|
TUBGCP5
|
tubulin, gamma complex associated protein 5 |
| chr6_-_32784687 | 0.42 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr11_-_67374177 | 0.41 |
ENST00000333139.3
|
C11orf72
|
chromosome 11 open reading frame 72 |
| chr9_-_113100088 | 0.41 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr12_-_133532864 | 0.41 |
ENST00000536932.1
ENST00000360187.4 ENST00000392321.3 |
CHFR
ZNF605
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase zinc finger protein 605 |
| chr4_+_40194570 | 0.40 |
ENST00000507851.1
|
RHOH
|
ras homolog family member H |
| chr19_-_56826157 | 0.40 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr16_-_89785777 | 0.39 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr11_-_61124266 | 0.38 |
ENST00000539890.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr19_+_44645700 | 0.38 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr9_-_70488865 | 0.37 |
ENST00000377392.5
|
CBWD5
|
COBW domain containing 5 |
| chr19_-_58220517 | 0.37 |
ENST00000512439.2
ENST00000426889.1 |
ZNF154
|
zinc finger protein 154 |
| chr11_-_115127611 | 0.37 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr19_+_56368803 | 0.37 |
ENST00000587891.1
|
NLRP4
|
NLR family, pyrin domain containing 4 |
| chr19_+_39881951 | 0.37 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr2_-_142888573 | 0.36 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr15_+_42066740 | 0.36 |
ENST00000514566.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr16_+_50914085 | 0.36 |
ENST00000576842.1
|
CTD-2034I21.2
|
CTD-2034I21.2 |
| chr4_+_25378912 | 0.35 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr8_-_56685859 | 0.35 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr19_-_36980455 | 0.34 |
ENST00000454319.1
ENST00000392170.2 |
ZNF566
|
zinc finger protein 566 |
| chr19_-_58427944 | 0.34 |
ENST00000312026.5
ENST00000536263.1 ENST00000598629.1 ENST00000595559.1 ENST00000597515.1 ENST00000599251.1 ENST00000598526.1 |
ZNF417
|
zinc finger protein 417 |
| chr5_-_177423243 | 0.33 |
ENST00000308304.2
|
PROP1
|
PROP paired-like homeobox 1 |
| chr20_+_54987168 | 0.33 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr7_+_142985308 | 0.33 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr2_+_238877424 | 0.33 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr5_+_178450753 | 0.33 |
ENST00000444149.2
ENST00000519896.1 ENST00000522442.1 |
ZNF879
|
zinc finger protein 879 |
| chr3_+_169940153 | 0.33 |
ENST00000295797.4
|
PRKCI
|
protein kinase C, iota |
| chr4_+_86396265 | 0.33 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_25378826 | 0.32 |
ENST00000315368.3
|
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr8_-_56685966 | 0.32 |
ENST00000334667.2
|
TMEM68
|
transmembrane protein 68 |
| chr3_-_183735731 | 0.31 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_-_37958086 | 0.31 |
ENST00000592490.1
ENST00000392149.2 |
ZNF569
|
zinc finger protein 569 |
| chr5_+_118668846 | 0.31 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_-_87564620 | 0.31 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr17_+_38097727 | 0.30 |
ENST00000377924.4
|
LRRC3C
|
leucine rich repeat containing 3C |
| chr5_+_32585605 | 0.30 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr5_+_68856035 | 0.30 |
ENST00000512736.1
ENST00000510979.1 ENST00000514162.1 ENST00000380729.3 ENST00000508344.2 |
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr8_+_27632083 | 0.29 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_+_57784826 | 0.29 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr4_+_130017268 | 0.29 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr19_+_37341260 | 0.29 |
ENST00000589046.1
|
ZNF345
|
zinc finger protein 345 |
| chr19_+_44764031 | 0.29 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr2_-_44223089 | 0.29 |
ENST00000447246.1
ENST00000409946.1 ENST00000409659.1 |
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr7_-_108096822 | 0.29 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT5B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.5 | 10.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.5 | 1.4 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.4 | 1.8 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 5.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 1.1 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.3 | 1.3 | GO:1990569 | GDP-fucose transport(GO:0015783) UDP-N-acetylglucosamine transport(GO:0015788) purine nucleotide-sugar transport(GO:0036079) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.2 | 3.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.4 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 0.6 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 0.5 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.2 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.5 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.6 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.7 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 0.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 1.7 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.7 | GO:0003095 | pressure natriuresis(GO:0003095) vitamin E metabolic process(GO:0042360) |
| 0.1 | 2.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 2.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 3.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 2.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.2 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.7 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 1.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 2.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.6 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.9 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 1.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 1.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 1.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 3.6 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 5.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.9 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.3 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 10.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 4.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.5 | 2.0 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.3 | 1.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.3 | 0.8 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.3 | 1.3 | GO:0005462 | GDP-fucose transmembrane transporter activity(GO:0005457) UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.2 | 1.8 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.2 | 0.8 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.5 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 4.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 1.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 2.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.5 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 1.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 9.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0015165 | nucleotide-sugar transmembrane transporter activity(GO:0005338) pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 3.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 3.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 24.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |