Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
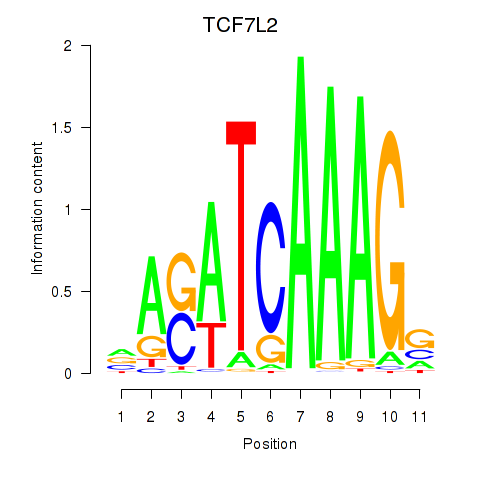
Results for TCF7L2
Z-value: 1.19
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.11 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg19_v2_chr10_+_114710516_114710675 | -0.11 | 5.3e-01 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_234621551 | 3.13 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr13_+_24144796 | 2.92 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr13_+_24144509 | 2.64 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_114128522 | 2.60 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr17_-_46035187 | 2.59 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr14_-_65409502 | 2.48 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr10_+_115312766 | 2.42 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr1_-_119869846 | 2.42 |
ENST00000457719.1
|
RP11-418J17.3
|
RP11-418J17.3 |
| chr17_+_68100989 | 2.39 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_+_69455855 | 2.35 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr9_+_97766469 | 2.21 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chrX_-_32173579 | 2.17 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr9_+_97766409 | 2.15 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr2_-_88427568 | 2.03 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr5_-_16936340 | 1.84 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr5_-_135290651 | 1.84 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr12_+_108168162 | 1.73 |
ENST00000342331.4
|
ASCL4
|
achaete-scute family bHLH transcription factor 4 |
| chr10_+_115312825 | 1.67 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr17_+_68101117 | 1.67 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_+_78400375 | 1.65 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr8_-_119964434 | 1.60 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chrX_+_46937745 | 1.60 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chrX_+_106163626 | 1.59 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr19_+_50084561 | 1.45 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr5_-_153857819 | 1.45 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr10_+_7745232 | 1.33 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr2_-_241195452 | 1.31 |
ENST00000457178.1
|
AC124861.1
|
AC124861.1 |
| chr13_+_76334795 | 1.21 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr13_+_76334567 | 1.17 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr1_-_219615984 | 1.15 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr16_+_72088376 | 1.10 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr17_+_42081914 | 1.05 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr3_+_29323043 | 1.04 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr14_+_68189190 | 1.01 |
ENST00000539142.1
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr15_+_43985725 | 1.00 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_+_7745303 | 1.00 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_101542462 | 0.98 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr7_+_73868439 | 0.96 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr17_+_79679299 | 0.96 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_-_134093275 | 0.96 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr1_-_110933611 | 0.95 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr13_+_76334498 | 0.94 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr18_+_19668021 | 0.93 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr1_-_110933663 | 0.92 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr3_-_134093395 | 0.92 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr6_-_112575912 | 0.92 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr1_-_94079648 | 0.91 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr15_+_43885252 | 0.90 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_+_152760397 | 0.89 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chr22_+_29279552 | 0.87 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr19_-_44008863 | 0.86 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr12_+_85673868 | 0.85 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr7_-_27213893 | 0.83 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr3_-_149375783 | 0.83 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr6_-_112575687 | 0.80 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr19_-_40791160 | 0.80 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_67138015 | 0.79 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr7_-_148725733 | 0.79 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr2_+_228678550 | 0.78 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chrX_+_2670066 | 0.77 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr2_-_171571077 | 0.76 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr1_+_78383813 | 0.76 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr5_+_131593364 | 0.75 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr6_+_32146131 | 0.74 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr3_+_174577070 | 0.74 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr20_+_57875658 | 0.74 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr1_+_162351503 | 0.73 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr2_+_69240302 | 0.71 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr15_+_43985084 | 0.71 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr17_+_1674982 | 0.71 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_94443065 | 0.69 |
ENST00000555287.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr6_-_112575758 | 0.67 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr6_-_11779840 | 0.65 |
ENST00000506810.1
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr9_-_27005686 | 0.65 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr7_-_148725544 | 0.64 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr4_-_65275100 | 0.64 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr14_-_94443105 | 0.63 |
ENST00000555019.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chrX_-_10588595 | 0.63 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr6_-_25930819 | 0.63 |
ENST00000360488.3
|
SLC17A2
|
solute carrier family 17, member 2 |
| chr5_-_16742330 | 0.63 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr11_-_76155618 | 0.63 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_61330931 | 0.63 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chrX_+_110754888 | 0.62 |
ENST00000569275.1
ENST00000563467.1 |
LINC00890
|
long intergenic non-protein coding RNA 890 |
| chr19_-_40791211 | 0.62 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr4_+_30721968 | 0.61 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr6_-_25930904 | 0.61 |
ENST00000377850.3
|
SLC17A2
|
solute carrier family 17, member 2 |
| chr8_-_124665190 | 0.60 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr4_+_87857538 | 0.60 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr7_-_16505440 | 0.60 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr16_-_73082274 | 0.58 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr19_-_40791302 | 0.57 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_171571827 | 0.57 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr13_+_102142296 | 0.57 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_+_69240415 | 0.56 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr3_-_155011483 | 0.56 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr7_+_137761167 | 0.56 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr8_+_55467072 | 0.56 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr2_+_177028805 | 0.56 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr10_-_75351088 | 0.54 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr17_-_79919713 | 0.54 |
ENST00000425009.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr4_-_146859787 | 0.54 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr2_+_69240511 | 0.53 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr11_-_108422926 | 0.52 |
ENST00000428840.1
ENST00000526312.1 |
EXPH5
|
exophilin 5 |
| chr10_+_54074033 | 0.52 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr20_-_656437 | 0.52 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr7_+_137761220 | 0.51 |
ENST00000242375.3
ENST00000438242.1 |
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr9_+_136399929 | 0.50 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr17_+_70026795 | 0.49 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr9_+_128510454 | 0.48 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr5_+_140235469 | 0.48 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr5_-_135290705 | 0.48 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_27219849 | 0.46 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_-_104192405 | 0.45 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr4_-_157892498 | 0.44 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr11_-_76155700 | 0.44 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr5_-_175815565 | 0.43 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr12_-_23737534 | 0.43 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr16_+_19222479 | 0.43 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr1_+_223101757 | 0.43 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr4_-_157892167 | 0.42 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr2_-_71357344 | 0.42 |
ENST00000494660.2
ENST00000244217.5 ENST00000486135.1 |
MCEE
|
methylmalonyl CoA epimerase |
| chr7_+_137761199 | 0.41 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr20_+_57875758 | 0.40 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr17_+_59529743 | 0.40 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr17_-_56065484 | 0.39 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr5_+_54455946 | 0.39 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr1_-_205419053 | 0.39 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr4_-_65275162 | 0.39 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr3_+_155838337 | 0.38 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_-_11907829 | 0.38 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr10_+_114709999 | 0.37 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_109935819 | 0.36 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr3_-_79068138 | 0.36 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_+_62417957 | 0.34 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr17_+_48611853 | 0.34 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr5_+_140797296 | 0.33 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr2_-_233641265 | 0.33 |
ENST00000438786.1
ENST00000409779.1 ENST00000233826.3 |
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr14_-_23762777 | 0.33 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr4_-_157892055 | 0.33 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_-_56065540 | 0.32 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr4_-_109684120 | 0.32 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr3_-_134093738 | 0.31 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr19_+_50003781 | 0.30 |
ENST00000602157.1
|
hsa-mir-150
|
hsa-mir-150 |
| chr7_-_25702558 | 0.30 |
ENST00000423689.2
|
AC003090.1
|
AC003090.1 |
| chrX_-_47004437 | 0.30 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr15_+_43885799 | 0.30 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_+_69488155 | 0.29 |
ENST00000374495.3
|
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr12_+_9067123 | 0.29 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr11_-_75921780 | 0.28 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr18_+_46065483 | 0.28 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr16_+_53242350 | 0.27 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr10_-_61900762 | 0.27 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chrX_+_54835493 | 0.27 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr14_+_62037287 | 0.27 |
ENST00000556569.1
|
RP11-47I22.3
|
Uncharacterized protein |
| chr11_-_6677018 | 0.27 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr1_-_54303934 | 0.27 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr2_+_48796120 | 0.26 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr21_-_35284635 | 0.26 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chrX_+_41306575 | 0.25 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr13_-_72440901 | 0.25 |
ENST00000359684.2
|
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr5_+_140734570 | 0.24 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr18_+_46065570 | 0.24 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr6_+_64281906 | 0.23 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_+_201936458 | 0.23 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chrX_-_47004878 | 0.23 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr8_+_75015699 | 0.23 |
ENST00000522498.1
|
RP11-6I2.4
|
RP11-6I2.4 |
| chr8_+_55466915 | 0.23 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chrX_+_69488174 | 0.23 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr1_+_200860122 | 0.23 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr4_-_146859623 | 0.22 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr20_+_57875457 | 0.22 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chrX_-_10588459 | 0.22 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr4_+_95972822 | 0.22 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr18_+_46065393 | 0.22 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr10_+_114710211 | 0.21 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr14_-_21502944 | 0.21 |
ENST00000382951.3
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active) |
| chr18_-_24445729 | 0.21 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr2_+_201936707 | 0.21 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr12_+_80603233 | 0.20 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr5_+_102201509 | 0.20 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_-_90294523 | 0.20 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chrX_+_15525426 | 0.20 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr12_-_130529501 | 0.20 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr3_-_45957534 | 0.20 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_+_57875711 | 0.19 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr6_-_112575838 | 0.19 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr8_-_142012169 | 0.19 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_154244987 | 0.18 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr7_+_27282319 | 0.18 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr10_+_114133773 | 0.18 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr7_+_80275621 | 0.17 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_180199393 | 0.17 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr1_+_147013182 | 0.17 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr1_-_54304212 | 0.17 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chrX_+_47004599 | 0.16 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr9_+_125796806 | 0.16 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr1_-_65533390 | 0.16 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr14_+_32798462 | 0.16 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_-_45957088 | 0.15 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr15_+_93447675 | 0.15 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.6 | 1.8 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.5 | 1.6 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.5 | 1.6 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.5 | 1.5 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.4 | 1.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.3 | 1.0 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.3 | 1.0 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.3 | 1.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.3 | 2.0 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.3 | 1.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 0.8 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.7 | GO:0100012 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.2 | 2.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.4 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.6 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 0.5 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 0.9 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 2.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.5 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.2 | 2.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.3 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 1.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 1.8 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 4.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.5 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.8 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 5.5 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 2.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.3 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 1.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 2.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 2.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 1.0 | GO:0045494 | retinol metabolic process(GO:0042572) photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 2.9 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 1.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.5 | GO:0050953 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.6 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.2 | 2.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 4.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 2.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 2.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 3.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 5.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.3 | 1.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.3 | 1.5 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.3 | 2.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 0.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 2.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 1.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.5 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 4.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 7.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 3.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 1.0 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 2.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |