Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for TP63
Z-value: 2.17
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189349162_189349305 | 0.52 | 2.5e-03 | Click! |
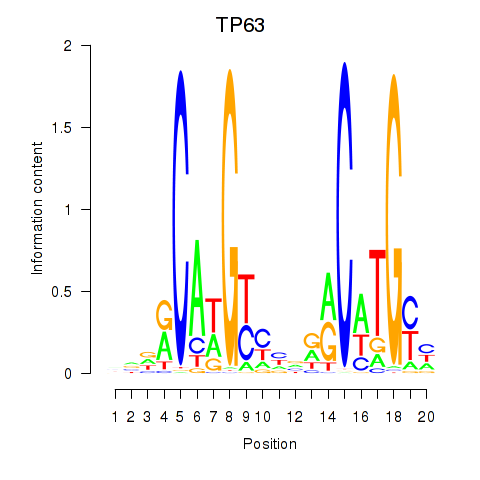
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_2670066 | 4.05 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chrX_+_2670153 | 3.21 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr7_+_80267949 | 2.91 |
ENST00000482059.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_18287721 | 2.76 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr11_+_18287801 | 2.74 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr19_+_18496957 | 2.71 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr20_-_52790512 | 2.55 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr1_+_17575584 | 2.54 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr1_-_209824643 | 2.45 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr9_+_124088860 | 2.45 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr7_+_80267973 | 2.45 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_-_42448788 | 2.45 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr10_-_88729200 | 2.38 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr9_+_35906176 | 2.37 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr11_-_111782484 | 2.36 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr6_-_46922659 | 2.34 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr6_+_54711533 | 2.34 |
ENST00000306858.7
|
FAM83B
|
family with sequence similarity 83, member B |
| chr11_-_111782696 | 2.32 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr10_-_88729069 | 2.31 |
ENST00000609457.1
|
MMRN2
|
multimerin 2 |
| chr1_-_153588334 | 2.28 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr11_+_393428 | 2.27 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr18_+_61143994 | 2.27 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr12_+_119616447 | 2.12 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr1_-_47655686 | 2.10 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr12_-_52887034 | 2.09 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr1_-_201398985 | 2.09 |
ENST00000336092.4
|
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr11_-_75921780 | 2.05 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr15_+_74466744 | 2.03 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr13_-_78492955 | 1.97 |
ENST00000446573.1
|
EDNRB
|
endothelin receptor type B |
| chr11_-_10920714 | 1.94 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr19_-_17375527 | 1.93 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr13_-_78492927 | 1.93 |
ENST00000334286.5
|
EDNRB
|
endothelin receptor type B |
| chr9_+_71944241 | 1.88 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr13_+_23755127 | 1.87 |
ENST00000545013.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_+_120031264 | 1.86 |
ENST00000426426.1
|
TMEM233
|
transmembrane protein 233 |
| chr10_-_90751038 | 1.84 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr14_+_51955831 | 1.82 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr3_+_132316081 | 1.81 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr15_-_89456630 | 1.80 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr12_-_52845910 | 1.79 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr13_+_23755054 | 1.78 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr15_-_89456593 | 1.77 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr2_+_234600253 | 1.76 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr11_+_111782934 | 1.76 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr11_+_111783450 | 1.74 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr19_+_35849362 | 1.74 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr3_+_138066539 | 1.72 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr8_+_33613265 | 1.72 |
ENST00000517292.1
|
RP11-317N12.1
|
RP11-317N12.1 |
| chr15_+_101417919 | 1.70 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr19_-_5293243 | 1.69 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr13_+_23755099 | 1.63 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_-_15942503 | 1.54 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr20_+_57264187 | 1.51 |
ENST00000525967.1
ENST00000525817.1 |
NPEPL1
|
aminopeptidase-like 1 |
| chr16_+_66413128 | 1.49 |
ENST00000563425.2
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr2_+_217524323 | 1.49 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_+_121756809 | 1.48 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr2_-_151344172 | 1.44 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr15_-_90233907 | 1.43 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr11_+_76493294 | 1.42 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr11_+_62009653 | 1.36 |
ENST00000244926.3
|
SCGB1D2
|
secretoglobin, family 1D, member 2 |
| chr1_-_117753540 | 1.36 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr5_-_169626104 | 1.35 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr11_+_832804 | 1.34 |
ENST00000397420.3
ENST00000525718.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr1_+_86889769 | 1.32 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr12_-_88974236 | 1.31 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr6_-_150212029 | 1.29 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr17_-_34079897 | 1.26 |
ENST00000254466.6
ENST00000587565.1 |
GAS2L2
|
growth arrest-specific 2 like 2 |
| chr19_+_35861831 | 1.26 |
ENST00000454971.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr17_+_21279509 | 1.26 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr16_+_82090028 | 1.24 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr7_-_72742085 | 1.23 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr16_-_67224002 | 1.23 |
ENST00000563889.1
ENST00000564418.1 ENST00000545725.2 ENST00000314586.6 |
EXOC3L1
|
exocyst complex component 3-like 1 |
| chr22_+_31489344 | 1.22 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr1_-_46089718 | 1.20 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr7_+_100466433 | 1.19 |
ENST00000429658.1
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr11_+_64073699 | 1.17 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_+_26485511 | 1.15 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chr3_+_69915385 | 1.12 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_165698662 | 1.12 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr16_+_67197288 | 1.12 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chrX_-_153602991 | 1.11 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr12_-_6233828 | 1.11 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr16_-_68000717 | 1.11 |
ENST00000541864.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr3_+_69915363 | 1.10 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_-_149375783 | 1.10 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_+_1856034 | 1.08 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr19_-_5292781 | 1.07 |
ENST00000586065.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr12_-_55042140 | 1.07 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr15_+_55700741 | 1.05 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr11_+_27062272 | 1.05 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr2_+_69201705 | 1.03 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr15_-_63448973 | 0.99 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr2_+_71127699 | 0.98 |
ENST00000234392.2
|
VAX2
|
ventral anterior homeobox 2 |
| chr12_-_96184533 | 0.98 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr19_+_35849723 | 0.98 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr15_+_41136216 | 0.98 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr3_-_50340996 | 0.96 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr4_-_145061788 | 0.95 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr19_+_15160130 | 0.94 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr16_+_19467772 | 0.94 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr20_-_6103666 | 0.94 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr4_+_15376165 | 0.93 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr12_+_52306113 | 0.93 |
ENST00000547400.1
ENST00000550683.1 ENST00000419526.2 |
ACVRL1
|
activin A receptor type II-like 1 |
| chr15_+_41136263 | 0.91 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr6_+_148663729 | 0.90 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr2_-_165698521 | 0.89 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr15_+_41136369 | 0.87 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chrX_-_153583257 | 0.87 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr20_+_61867235 | 0.85 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr7_-_1600433 | 0.84 |
ENST00000431208.1
|
TMEM184A
|
transmembrane protein 184A |
| chr1_-_94312706 | 0.83 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_118727781 | 0.83 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr5_-_156486120 | 0.82 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr1_+_152799949 | 0.82 |
ENST00000335123.2
|
LCE1A
|
late cornified envelope 1A |
| chr1_-_19229014 | 0.82 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr8_+_74206829 | 0.82 |
ENST00000240285.5
|
RDH10
|
retinol dehydrogenase 10 (all-trans) |
| chr19_-_17375541 | 0.81 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr19_-_4717835 | 0.81 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr3_+_32148106 | 0.78 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr11_+_1049862 | 0.78 |
ENST00000534584.1
|
RP13-870H17.3
|
RP13-870H17.3 |
| chr8_-_49833978 | 0.77 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr7_+_102389434 | 0.77 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr14_-_23904861 | 0.75 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr1_-_19229248 | 0.74 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr16_-_57837129 | 0.74 |
ENST00000562984.1
ENST00000564891.1 |
KIFC3
|
kinesin family member C3 |
| chr4_+_186317133 | 0.73 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr2_-_96812751 | 0.73 |
ENST00000449242.1
|
AC012307.2
|
AC012307.2 |
| chr14_+_24779340 | 0.72 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr5_-_42812143 | 0.72 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_8160416 | 0.72 |
ENST00000505872.1
ENST00000447017.2 ENST00000341937.5 ENST00000361581.5 |
ABLIM2
|
actin binding LIM protein family, member 2 |
| chr19_+_35862192 | 0.71 |
ENST00000597214.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr12_-_96184913 | 0.71 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr19_-_47288162 | 0.69 |
ENST00000594991.1
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr7_+_79763271 | 0.69 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr4_-_87374330 | 0.69 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_134210243 | 0.69 |
ENST00000367882.4
|
TCF21
|
transcription factor 21 |
| chr1_+_236558694 | 0.68 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr15_-_90233866 | 0.68 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr16_+_57653625 | 0.68 |
ENST00000567553.1
ENST00000565314.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57653596 | 0.68 |
ENST00000568791.1
ENST00000570044.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr4_-_129209221 | 0.66 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr19_-_19051103 | 0.66 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr15_+_86686953 | 0.65 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr6_+_18387570 | 0.63 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr20_+_34679725 | 0.63 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr18_+_34124507 | 0.62 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr8_-_74206133 | 0.62 |
ENST00000352983.2
|
RPL7
|
ribosomal protein L7 |
| chr22_+_36044411 | 0.61 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr3_+_136676851 | 0.61 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr20_+_48807351 | 0.61 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr19_-_6591113 | 0.61 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr15_+_69857515 | 0.61 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_-_19229218 | 0.60 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr19_+_12175535 | 0.60 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr3_+_136676707 | 0.60 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr12_-_58159361 | 0.60 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chrX_+_152086373 | 0.60 |
ENST00000318529.8
|
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr17_-_74707037 | 0.58 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr14_-_46185155 | 0.58 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr2_+_201987200 | 0.58 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr7_-_100287071 | 0.57 |
ENST00000275732.5
|
GIGYF1
|
GRB10 interacting GYF protein 1 |
| chr16_-_57798253 | 0.57 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr1_-_3528034 | 0.57 |
ENST00000356575.4
|
MEGF6
|
multiple EGF-like-domains 6 |
| chr19_+_12175596 | 0.57 |
ENST00000441304.2
|
ZNF844
|
zinc finger protein 844 |
| chr19_-_11688951 | 0.56 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr20_-_43743790 | 0.56 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr19_-_11639931 | 0.55 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr19_-_11688500 | 0.55 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr5_+_162864575 | 0.55 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr9_+_139553306 | 0.55 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr1_-_26197744 | 0.55 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr4_+_72204755 | 0.54 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr9_+_101569944 | 0.54 |
ENST00000375011.3
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr21_+_44313375 | 0.53 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr18_+_61144160 | 0.53 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr2_-_31361362 | 0.53 |
ENST00000430167.1
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr11_-_67397371 | 0.52 |
ENST00000376693.2
ENST00000301490.4 |
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr7_-_3197888 | 0.52 |
ENST00000458648.1
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr20_-_23669590 | 0.51 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chr11_+_47608198 | 0.51 |
ENST00000356737.2
ENST00000538490.1 |
FAM180B
|
family with sequence similarity 180, member B |
| chr12_-_49488573 | 0.50 |
ENST00000266991.2
|
DHH
|
desert hedgehog |
| chr20_+_34894247 | 0.50 |
ENST00000373913.3
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr14_-_37642016 | 0.50 |
ENST00000331299.5
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr5_-_160365601 | 0.50 |
ENST00000518580.1
ENST00000518301.1 |
RP11-109J4.1
|
RP11-109J4.1 |
| chr4_-_10023095 | 0.49 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr7_+_75024903 | 0.49 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr3_+_37441062 | 0.49 |
ENST00000426078.1
|
C3orf35
|
chromosome 3 open reading frame 35 |
| chr17_-_28661065 | 0.47 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr5_-_180235755 | 0.47 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr5_-_42811986 | 0.47 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr12_+_120105558 | 0.46 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr15_-_55700522 | 0.46 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr13_-_96296944 | 0.46 |
ENST00000361396.2
ENST00000376829.2 |
DZIP1
|
DAZ interacting zinc finger protein 1 |
| chr14_-_37641618 | 0.45 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr2_-_31361543 | 0.44 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr22_+_23040274 | 0.44 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr7_-_3198204 | 0.44 |
ENST00000402115.1
ENST00000403226.1 |
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr17_-_4938712 | 0.43 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr19_+_45165545 | 0.43 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr16_+_56995762 | 0.42 |
ENST00000200676.3
ENST00000379780.2 |
CETP
|
cholesteryl ester transfer protein, plasma |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0007497 | posterior midgut development(GO:0007497) endothelin receptor signaling pathway(GO:0086100) |
| 0.9 | 2.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.8 | 3.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.7 | 2.7 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.7 | 2.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.5 | 2.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.5 | 2.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 2.5 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.4 | 5.4 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.4 | 1.3 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.4 | 1.7 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.4 | 1.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 4.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 1.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.4 | 1.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.3 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 1.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.3 | 1.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 2.1 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.3 | 0.8 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 0.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 1.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.7 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.2 | 2.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 1.0 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 4.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.7 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 0.7 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 0.8 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 3.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.9 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.2 | 2.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.4 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 1.0 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 2.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 2.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 5.1 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 3.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 2.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 0.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) |
| 0.1 | 0.4 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 2.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 2.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 1.0 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.4 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 2.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.6 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.0 | 0.3 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 1.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 2.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.4 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.7 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 1.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 1.3 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 1.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0035936 | androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 5.6 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 1.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 1.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 0.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.3 | 4.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 2.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 5.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.9 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 4.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 5.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 6.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 3.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 2.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 2.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.5 | 2.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.5 | 2.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 5.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 2.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 0.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 1.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.2 | 3.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 1.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 2.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 2.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 4.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.4 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 1.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 5.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0004583 | alpha-1,3-mannosyltransferase activity(GO:0000033) dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 2.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 1.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 6.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 1.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.3 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.7 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 6.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 2.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 2.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.0 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 2.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 1.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 8.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 3.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 4.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 3.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 6.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 9.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 5.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 3.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.3 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 6.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |