Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
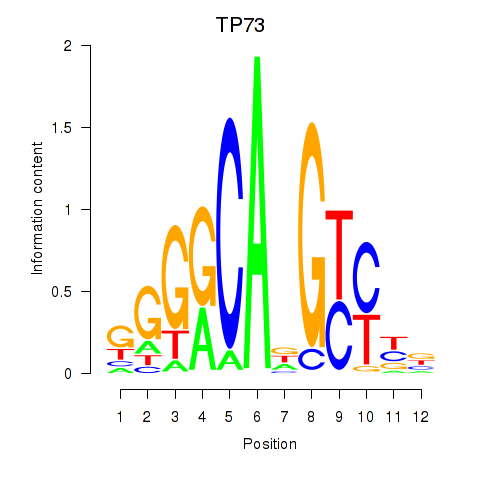
Results for TP73
Z-value: 1.26
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg19_v2_chr1_+_3598871_3598930 | 0.42 | 1.6e-02 | Click! |
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_222846 | 2.81 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr7_-_27213893 | 2.73 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr14_+_22217447 | 2.45 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr14_-_25045446 | 2.19 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chr7_-_128045984 | 1.97 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr7_-_100171270 | 1.95 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr14_+_75746340 | 1.91 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr15_+_75498355 | 1.75 |
ENST00000567617.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr4_-_87770416 | 1.72 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr15_+_41136216 | 1.70 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr15_+_75498739 | 1.63 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr2_+_128293323 | 1.58 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr14_+_75745477 | 1.55 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr14_+_22554680 | 1.50 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr2_+_114163945 | 1.47 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr17_-_42452063 | 1.46 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr14_+_95982451 | 1.44 |
ENST00000554161.1
|
RP11-1070N10.3
|
Uncharacterized protein |
| chr14_+_52780998 | 1.40 |
ENST00000557436.1
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr15_+_41136586 | 1.32 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr6_-_46703069 | 1.29 |
ENST00000538237.1
ENST00000274793.7 |
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr15_+_41136369 | 1.27 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr15_+_74466744 | 1.21 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr17_+_43318434 | 1.21 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr15_+_41136263 | 1.13 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_+_37356586 | 1.09 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr17_+_37356528 | 1.06 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr10_-_105212141 | 1.00 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr12_-_57505121 | 1.00 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr10_-_105212059 | 0.99 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr19_+_45254529 | 0.99 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr14_-_75078725 | 0.97 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr3_-_185538849 | 0.96 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_+_47137445 | 0.96 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr1_-_160832490 | 0.95 |
ENST00000322302.7
ENST00000368033.3 |
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr2_-_26864228 | 0.94 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr17_+_37356555 | 0.93 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr1_-_160832642 | 0.92 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr6_-_46703430 | 0.91 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr22_-_37976082 | 0.91 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr9_-_130517309 | 0.90 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C |
| chr8_+_37654693 | 0.88 |
ENST00000412232.2
|
GPR124
|
G protein-coupled receptor 124 |
| chr12_+_102271436 | 0.88 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_+_47137544 | 0.87 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr3_+_136676851 | 0.83 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr9_+_131445703 | 0.83 |
ENST00000454747.1
|
SET
|
SET nuclear oncogene |
| chr11_+_65154070 | 0.81 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr8_+_37654424 | 0.80 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr20_+_35504522 | 0.79 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr3_+_136676707 | 0.77 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr10_+_47746929 | 0.75 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr17_+_73750699 | 0.75 |
ENST00000584939.1
|
ITGB4
|
integrin, beta 4 |
| chr17_+_54230819 | 0.74 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr17_+_7792101 | 0.74 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr20_+_48807351 | 0.72 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr9_-_20382446 | 0.72 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_-_112194484 | 0.69 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr14_-_36983034 | 0.69 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr9_+_6758024 | 0.68 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr18_-_53253323 | 0.68 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr1_-_160832670 | 0.66 |
ENST00000368032.2
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr17_-_42200996 | 0.66 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr17_-_80059726 | 0.62 |
ENST00000583053.1
|
CCDC57
|
coiled-coil domain containing 57 |
| chr11_+_75479850 | 0.61 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chrX_-_38186811 | 0.60 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr19_-_35981358 | 0.58 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr9_+_6757634 | 0.56 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr14_-_105531759 | 0.55 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr9_+_6758109 | 0.53 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_-_26197744 | 0.53 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr20_-_62203808 | 0.53 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr1_-_236228417 | 0.53 |
ENST00000264187.6
|
NID1
|
nidogen 1 |
| chr14_+_23006547 | 0.52 |
ENST00000390530.1
|
TRAJ7
|
T cell receptor alpha joining 7 |
| chr16_+_8891670 | 0.48 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr9_+_36169380 | 0.47 |
ENST00000335119.2
|
CCIN
|
calicin |
| chr2_+_220283091 | 0.47 |
ENST00000373960.3
|
DES
|
desmin |
| chr1_-_236228403 | 0.47 |
ENST00000366595.3
|
NID1
|
nidogen 1 |
| chr20_-_49253425 | 0.46 |
ENST00000045083.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr10_-_47173994 | 0.46 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr4_+_6717842 | 0.45 |
ENST00000320776.3
|
BLOC1S4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr2_+_95940186 | 0.43 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chrX_+_77166172 | 0.43 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr10_+_70661014 | 0.42 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr18_-_53253000 | 0.42 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr9_+_133223932 | 0.41 |
ENST00000420499.2
|
HMCN2
|
hemicentin 2 |
| chr19_+_41856816 | 0.40 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr17_-_40835076 | 0.39 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr19_+_45281118 | 0.39 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr10_-_79686284 | 0.38 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr5_-_156593273 | 0.38 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71, member B |
| chrX_-_38186775 | 0.37 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr10_-_99185798 | 0.35 |
ENST00000439965.2
|
AL355490.1
|
LOC644215 protein; Uncharacterized protein |
| chr18_-_53253112 | 0.35 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr15_+_64680003 | 0.34 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr19_+_44037546 | 0.34 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr22_+_39966758 | 0.34 |
ENST00000407673.1
ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr20_-_52790512 | 0.33 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr16_+_84178874 | 0.33 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_-_78444776 | 0.33 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr19_-_19626838 | 0.30 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr13_-_48877795 | 0.30 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr19_-_39226045 | 0.30 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr5_-_141060389 | 0.30 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr2_-_232328867 | 0.28 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr22_+_29168652 | 0.28 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr19_+_41620335 | 0.28 |
ENST00000331105.2
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1 |
| chr5_+_78532003 | 0.28 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr20_-_52790055 | 0.27 |
ENST00000395955.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr1_-_78444738 | 0.27 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr11_+_60197040 | 0.27 |
ENST00000300190.2
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_+_95940220 | 0.24 |
ENST00000542147.1
|
PROM2
|
prominin 2 |
| chr7_-_2883928 | 0.24 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr5_+_38258511 | 0.24 |
ENST00000354891.3
ENST00000322350.5 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr3_-_151160938 | 0.23 |
ENST00000489791.1
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr11_-_75062829 | 0.23 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr6_+_35995552 | 0.23 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr5_+_119799927 | 0.22 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr1_-_241799232 | 0.21 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr6_+_35995531 | 0.19 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr21_+_43442100 | 0.19 |
ENST00000455701.1
ENST00000596595.1 |
ZNF295-AS1
|
ZNF295 antisense RNA 1 |
| chr7_-_2883650 | 0.18 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr11_+_65686952 | 0.18 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr16_-_68000717 | 0.17 |
ENST00000541864.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr9_-_128003606 | 0.17 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr17_-_39023462 | 0.17 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr6_-_33256664 | 0.17 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr7_+_141618617 | 0.16 |
ENST00000548136.1
|
OR9A4
|
olfactory receptor, family 9, subfamily A, member 4 |
| chr16_+_67197288 | 0.15 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr8_-_70745575 | 0.14 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr16_+_67195592 | 0.14 |
ENST00000519378.1
|
FBXL8
|
F-box and leucine-rich repeat protein 8 |
| chr11_+_60197069 | 0.14 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr13_+_48877895 | 0.13 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr12_-_117319236 | 0.13 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr7_+_142636603 | 0.13 |
ENST00000409607.3
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr16_+_84178850 | 0.12 |
ENST00000334315.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr11_+_65687158 | 0.11 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr8_+_145065521 | 0.10 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_+_236540 | 0.10 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr7_+_142636440 | 0.10 |
ENST00000458732.1
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr12_-_82752159 | 0.09 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr14_+_72399833 | 0.09 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr17_+_78388959 | 0.09 |
ENST00000518137.1
ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV
|
endonuclease V |
| chr18_+_77623668 | 0.08 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr11_-_65686496 | 0.07 |
ENST00000449692.3
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr4_-_76598544 | 0.06 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrX_+_72667090 | 0.05 |
ENST00000373514.2
|
CDX4
|
caudal type homeobox 4 |
| chr17_+_4835580 | 0.05 |
ENST00000329125.5
|
GP1BA
|
glycoprotein Ib (platelet), alpha polypeptide |
| chr16_-_2581409 | 0.04 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
| chr12_+_113495492 | 0.04 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr9_+_131445928 | 0.04 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr2_-_27357479 | 0.03 |
ENST00000406567.3
ENST00000260643.2 |
PREB
|
prolactin regulatory element binding |
| chr12_+_44152740 | 0.02 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr1_+_155146318 | 0.02 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr5_+_138940742 | 0.02 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr5_+_43120985 | 0.01 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr17_-_28661065 | 0.01 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr17_-_47492236 | 0.00 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.4 | 1.6 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.4 | 3.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.3 | 2.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.3 | 2.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 2.2 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.2 | 5.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 1.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 0.7 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 0.6 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.6 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 3.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 2.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 1.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 1.4 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.7 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.9 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 1.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.5 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 1.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 0.5 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.7 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 2.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 4.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 2.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 1.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 2.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 1.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.9 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 3.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 3.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |