Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
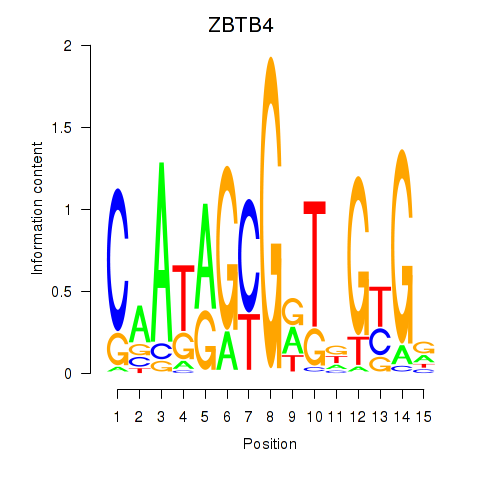
Results for ZBTB4
Z-value: 1.35
Transcription factors associated with ZBTB4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB4
|
ENSG00000174282.7 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB4 | hg19_v2_chr17_-_7387524_7387597 | -0.23 | 2.1e-01 | Click! |
Activity profile of ZBTB4 motif
Sorted Z-values of ZBTB4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_11370330 | 2.63 |
ENST00000241808.4
ENST00000435245.2 |
PRM2
|
protamine 2 |
| chr2_+_192109911 | 2.57 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr8_-_105479270 | 2.53 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr1_+_196912902 | 2.49 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr2_+_192110199 | 2.45 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr20_+_3052264 | 2.41 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr1_+_196788887 | 2.32 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr19_+_35596873 | 2.31 |
ENST00000313865.6
|
AC020907.1
|
Uncharacterized protein |
| chr11_-_1016149 | 2.27 |
ENST00000532016.1
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr11_-_72385437 | 2.24 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr3_-_128185811 | 2.04 |
ENST00000469083.1
|
DNAJB8
|
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chr14_+_85996471 | 1.91 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_34058639 | 1.89 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr17_-_19265855 | 1.87 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr6_+_41606176 | 1.85 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr7_+_87257701 | 1.85 |
ENST00000338056.3
ENST00000493037.1 |
RUNDC3B
|
RUN domain containing 3B |
| chr3_+_159943362 | 1.84 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr5_-_146889619 | 1.72 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr11_+_705193 | 1.40 |
ENST00000527199.1
|
EPS8L2
|
EPS8-like 2 |
| chr16_+_30996502 | 1.36 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_65886326 | 1.35 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr12_+_5541267 | 1.33 |
ENST00000423158.3
|
NTF3
|
neurotrophin 3 |
| chr14_+_21569245 | 1.33 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr6_-_49430886 | 1.31 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr8_-_27468945 | 1.30 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr8_-_27469196 | 1.27 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr10_+_17851362 | 1.22 |
ENST00000331429.2
ENST00000457317.1 |
MRC1L1
|
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr14_+_85996507 | 1.20 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr8_-_27468842 | 1.19 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr15_-_40212363 | 1.18 |
ENST00000299092.3
|
GPR176
|
G protein-coupled receptor 176 |
| chr19_+_50433476 | 1.17 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr1_+_65886244 | 1.17 |
ENST00000344610.8
|
LEPR
|
leptin receptor |
| chr15_+_90792760 | 1.15 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr22_+_29279552 | 1.12 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr22_+_21987005 | 1.12 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr16_-_75301886 | 1.09 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr7_+_87257854 | 1.07 |
ENST00000394654.3
|
RUNDC3B
|
RUN domain containing 3B |
| chr8_-_27468717 | 1.05 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chrX_-_99665262 | 1.04 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr17_-_30186328 | 1.04 |
ENST00000302362.6
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr17_-_30185971 | 1.03 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr2_-_179343226 | 1.02 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr13_-_21348050 | 1.00 |
ENST00000382754.4
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr16_+_66413128 | 0.99 |
ENST00000563425.2
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr13_-_21348066 | 0.98 |
ENST00000382758.1
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr8_-_27469383 | 0.97 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr16_-_30042580 | 0.96 |
ENST00000380495.4
|
FAM57B
|
family with sequence similarity 57, member B |
| chr19_+_6135646 | 0.96 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr6_-_136847610 | 0.96 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr8_-_143851203 | 0.95 |
ENST00000521396.1
ENST00000317543.7 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr9_-_93405352 | 0.95 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr3_-_128186091 | 0.93 |
ENST00000319153.3
|
DNAJB8
|
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chr9_-_80263220 | 0.91 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr19_-_17356697 | 0.91 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr22_+_32439019 | 0.89 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr9_+_70971815 | 0.89 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr9_-_71629010 | 0.89 |
ENST00000377276.2
|
PRKACG
|
protein kinase, cAMP-dependent, catalytic, gamma |
| chr8_-_71581377 | 0.89 |
ENST00000276590.4
ENST00000522447.1 |
LACTB2
|
lactamase, beta 2 |
| chr7_-_18067478 | 0.89 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr2_+_74685413 | 0.89 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr5_-_150948414 | 0.88 |
ENST00000261800.5
|
FAT2
|
FAT atypical cadherin 2 |
| chr14_+_23305760 | 0.88 |
ENST00000311852.6
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr2_+_74685527 | 0.87 |
ENST00000393972.3
ENST00000409737.1 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr3_-_57583130 | 0.86 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr1_+_169075554 | 0.86 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr20_-_36793774 | 0.85 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chrX_-_129519511 | 0.84 |
ENST00000276218.2
|
GPR119
|
G protein-coupled receptor 119 |
| chr8_-_74205851 | 0.84 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr3_+_188889737 | 0.80 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr13_-_33780133 | 0.80 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr14_-_23446900 | 0.79 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr3_-_57583052 | 0.78 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr19_+_19144384 | 0.78 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr12_-_120805872 | 0.77 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr3_+_148583043 | 0.77 |
ENST00000296046.3
|
CPA3
|
carboxypeptidase A3 (mast cell) |
| chr19_-_19144243 | 0.77 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr17_-_15165854 | 0.75 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr15_-_40213080 | 0.74 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr20_-_36793663 | 0.74 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr14_-_69446034 | 0.73 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr19_+_36024310 | 0.72 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr22_+_50312316 | 0.71 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr10_+_37414785 | 0.69 |
ENST00000374660.1
ENST00000602533.1 ENST00000361713.1 |
ANKRD30A
|
ankyrin repeat domain 30A |
| chr22_+_50312379 | 0.69 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chrX_-_71526813 | 0.68 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr14_-_69445968 | 0.67 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr11_+_1093318 | 0.67 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr14_-_23822080 | 0.67 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr7_+_30811004 | 0.67 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr10_+_123922941 | 0.66 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_-_30185946 | 0.64 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr4_-_113437328 | 0.64 |
ENST00000313341.3
|
NEUROG2
|
neurogenin 2 |
| chr11_+_1092184 | 0.64 |
ENST00000361558.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr14_+_23305783 | 0.63 |
ENST00000547279.1
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr11_-_64546202 | 0.61 |
ENST00000377390.3
ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1
|
splicing factor 1 |
| chr19_-_12476443 | 0.61 |
ENST00000242804.4
ENST00000438182.1 |
ZNF442
|
zinc finger protein 442 |
| chr4_+_183370146 | 0.61 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr10_+_18098352 | 0.60 |
ENST00000239761.3
|
MRC1
|
mannose receptor, C type 1 |
| chr14_-_74551096 | 0.59 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_+_154947126 | 0.58 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr6_+_159290917 | 0.58 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr9_-_131709858 | 0.57 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr12_+_57388230 | 0.57 |
ENST00000300098.1
|
GPR182
|
G protein-coupled receptor 182 |
| chr5_+_173416162 | 0.57 |
ENST00000340147.6
|
C5orf47
|
chromosome 5 open reading frame 47 |
| chr14_+_74416989 | 0.57 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr1_+_33219592 | 0.56 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr4_-_156297949 | 0.56 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr19_-_49865639 | 0.54 |
ENST00000593945.1
ENST00000601519.1 ENST00000539846.1 ENST00000596757.1 ENST00000311227.2 |
TEAD2
|
TEA domain family member 2 |
| chr1_+_154947148 | 0.54 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr12_-_91348949 | 0.53 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr11_-_46940074 | 0.52 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr15_+_42696992 | 0.52 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr17_-_56595196 | 0.50 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr16_+_85391069 | 0.49 |
ENST00000568080.1
|
RP11-680G10.1
|
Uncharacterized protein |
| chr3_+_187957646 | 0.49 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_-_80084594 | 0.48 |
ENST00000548426.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr10_-_35104185 | 0.48 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr19_-_18902106 | 0.47 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr5_-_137090028 | 0.47 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr7_+_100136811 | 0.46 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr9_-_16215897 | 0.45 |
ENST00000433347.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr1_-_203055129 | 0.45 |
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4) |
| chr3_-_19988462 | 0.44 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr19_+_1275917 | 0.44 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr19_+_15121532 | 0.44 |
ENST00000292574.3
|
CCDC105
|
coiled-coil domain containing 105 |
| chr14_-_69445793 | 0.44 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr17_+_7487146 | 0.43 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr12_+_112204691 | 0.43 |
ENST00000416293.3
ENST00000261733.2 |
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr18_+_12658185 | 0.43 |
ENST00000400514.2
|
AP005482.1
|
Uncharacterized protein |
| chr2_+_131331904 | 0.42 |
ENST00000440359.1
|
AC140481.2
|
Uncharacterized protein |
| chr18_+_12658209 | 0.41 |
ENST00000400512.1
|
AP005482.1
|
Uncharacterized protein |
| chr17_-_9479128 | 0.41 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr20_-_43438912 | 0.40 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr4_+_128554081 | 0.39 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr2_-_10952922 | 0.39 |
ENST00000272227.3
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr17_-_15165825 | 0.39 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr1_-_165738085 | 0.39 |
ENST00000464650.1
ENST00000392129.6 |
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr2_-_10952832 | 0.38 |
ENST00000540494.1
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr12_+_6561190 | 0.37 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr9_-_136242956 | 0.37 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr4_-_156297919 | 0.37 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr20_+_61147651 | 0.36 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr18_-_47340297 | 0.36 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr11_+_125462690 | 0.35 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr1_-_165738072 | 0.35 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr1_+_10534944 | 0.34 |
ENST00000356607.4
ENST00000538836.1 ENST00000491661.2 |
PEX14
|
peroxisomal biogenesis factor 14 |
| chrM_+_3299 | 0.34 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr17_-_40264321 | 0.33 |
ENST00000430773.1
ENST00000413196.2 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr9_+_131181798 | 0.33 |
ENST00000420512.1
|
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr15_+_91498089 | 0.33 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr8_-_145691031 | 0.32 |
ENST00000424149.2
ENST00000530637.1 ENST00000306145.5 |
CYHR1
|
cysteine/histidine-rich 1 |
| chr17_-_40264692 | 0.31 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr11_+_86511569 | 0.31 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr6_+_158244223 | 0.31 |
ENST00000392185.3
|
SNX9
|
sorting nexin 9 |
| chr14_+_74417192 | 0.30 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr13_-_50018140 | 0.29 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr3_+_113666748 | 0.29 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr1_-_36615051 | 0.28 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr15_+_42696954 | 0.28 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr19_+_47759716 | 0.28 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr16_-_79634595 | 0.28 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr11_+_86511549 | 0.27 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr10_+_105036909 | 0.26 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr1_-_67896009 | 0.25 |
ENST00000370990.5
|
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr16_-_30538079 | 0.25 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr6_+_49431073 | 0.25 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr19_-_39421377 | 0.25 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr1_+_110754094 | 0.24 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr8_-_94928861 | 0.23 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2 |
| chr21_-_38445470 | 0.23 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr17_+_4046964 | 0.22 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr7_+_102937869 | 0.22 |
ENST00000249269.4
ENST00000428154.1 ENST00000420236.2 |
PMPCB
|
peptidase (mitochondrial processing) beta |
| chr14_-_25479811 | 0.22 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr1_-_67896069 | 0.20 |
ENST00000370995.2
ENST00000361219.6 |
SERBP1
|
SERPINE1 mRNA binding protein 1 |
| chr5_-_115177247 | 0.20 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr1_-_36615065 | 0.20 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chrX_+_153237740 | 0.19 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr15_+_42697018 | 0.18 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr3_+_46742823 | 0.18 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr21_-_38445011 | 0.17 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr19_-_39881669 | 0.17 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_+_50354430 | 0.17 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr15_-_77363513 | 0.17 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr11_-_63439013 | 0.17 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chrX_-_74145273 | 0.17 |
ENST00000055682.6
|
KIAA2022
|
KIAA2022 |
| chrX_+_47867194 | 0.16 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr10_+_76586348 | 0.16 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr19_-_5791215 | 0.16 |
ENST00000320699.8
ENST00000309061.7 |
DUS3L
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr4_-_83295103 | 0.16 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr9_-_111696340 | 0.15 |
ENST00000374647.5
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr1_+_43148059 | 0.14 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr12_+_52281744 | 0.14 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chrX_-_68385274 | 0.14 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr18_-_12657988 | 0.13 |
ENST00000410092.3
ENST00000409402.4 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr1_+_43148625 | 0.13 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr10_+_35416223 | 0.13 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr2_+_74361599 | 0.13 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr9_+_111696664 | 0.13 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr22_-_19929321 | 0.13 |
ENST00000400519.1
ENST00000535882.1 ENST00000334363.9 ENST00000400521.1 |
TXNRD2
|
thioredoxin reductase 2 |
| chr4_+_71570430 | 0.12 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.8 | 3.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.7 | 5.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 1.9 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.5 | 3.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 4.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 2.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.3 | 1.6 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 1.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 2.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 2.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.2 | 0.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 1.5 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 1.0 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.7 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.4 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 3.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.3 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 1.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.9 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 3.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.4 | GO:0014735 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.7 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 | 1.0 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 1.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.6 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:1904585 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 2.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.9 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.8 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.9 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.9 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.5 | GO:0014877 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 4.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.1 | GO:0048011 | cellular response to hepatocyte growth factor stimulus(GO:0035729) neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.7 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 1.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.0 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 1.9 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 1.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.3 | 5.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 1.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0002054 | nucleobase binding(GO:0002054) dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 2.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 5.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 1.9 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 5.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 2.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 1.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 3.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 2.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.0 | GO:0047617 | very long-chain fatty acid-CoA ligase activity(GO:0031957) acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 5.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 3.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 4.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |