Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
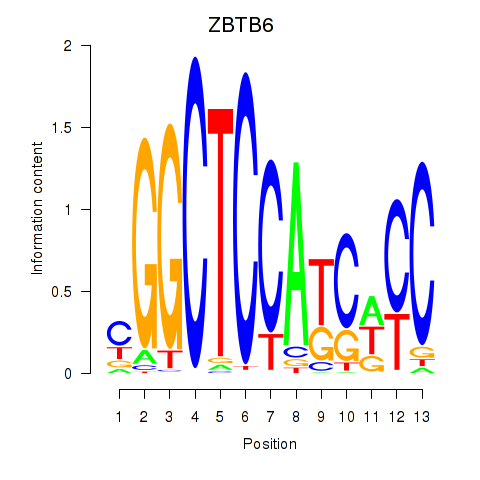
Results for ZBTB6
Z-value: 2.65
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.4 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg19_v2_chr9_-_125675576_125675612 | 0.41 | 2.1e-02 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_104513086 | 6.05 |
ENST00000406091.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_+_24540154 | 5.31 |
ENST00000559778.1
ENST00000560761.1 ENST00000557889.1 |
CPNE6
|
copine VI (neuronal) |
| chr12_-_6809543 | 5.12 |
ENST00000540656.1
|
PIANP
|
PILR alpha associated neural protein |
| chr12_-_50298000 | 4.78 |
ENST00000550635.2
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr9_+_34958254 | 4.70 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr3_-_116164306 | 4.44 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_+_110693103 | 4.31 |
ENST00000331565.4
|
SLC6A17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr3_+_181429704 | 4.13 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr14_+_24540731 | 4.11 |
ENST00000558859.1
ENST00000559197.1 ENST00000560828.1 ENST00000216775.2 ENST00000560884.1 |
CPNE6
|
copine VI (neuronal) |
| chr2_-_220174166 | 4.06 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr12_-_6809958 | 4.05 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr4_+_159131630 | 4.05 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr6_+_44238203 | 4.03 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr4_+_159131346 | 3.94 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr11_-_12030905 | 3.94 |
ENST00000326932.4
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr3_-_116163830 | 3.89 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_-_160040038 | 3.87 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr2_+_220492373 | 3.81 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr4_+_159131596 | 3.71 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr2_+_220492287 | 3.67 |
ENST00000273063.6
ENST00000373762.3 |
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_-_39239987 | 3.64 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr16_+_2039946 | 3.41 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr5_+_169931009 | 3.36 |
ENST00000328939.4
ENST00000390656.4 |
KCNIP1
|
Kv channel interacting protein 1 |
| chr4_-_163085141 | 3.32 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr2_+_166095898 | 3.30 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr5_-_149682447 | 3.26 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr9_+_87284622 | 3.24 |
ENST00000395882.1
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_104512976 | 3.23 |
ENST00000504942.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_2061119 | 3.23 |
ENST00000423729.2
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative) |
| chr11_+_6260298 | 3.22 |
ENST00000379936.2
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr4_-_87278857 | 3.18 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_+_4304585 | 3.18 |
ENST00000221856.6
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr2_-_193059634 | 3.17 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr19_+_4304632 | 3.17 |
ENST00000597590.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr2_+_220491973 | 3.15 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr18_+_48086440 | 3.14 |
ENST00000400384.2
ENST00000540640.1 ENST00000592595.1 |
MAPK4
|
mitogen-activated protein kinase 4 |
| chr2_-_135476552 | 3.13 |
ENST00000281924.6
|
TMEM163
|
transmembrane protein 163 |
| chr2_+_220492116 | 3.12 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_+_159943362 | 3.10 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr10_-_108924284 | 3.04 |
ENST00000344440.6
ENST00000263054.6 |
SORCS1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr11_-_12030746 | 3.00 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr3_-_133614421 | 2.93 |
ENST00000543906.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr9_+_87284675 | 2.91 |
ENST00000376208.1
ENST00000304053.6 ENST00000277120.3 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_-_102582155 | 2.89 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr6_-_166721871 | 2.86 |
ENST00000322583.3
|
PRR18
|
proline rich 18 |
| chr17_+_7608511 | 2.81 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr3_+_10857885 | 2.81 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr4_-_163085107 | 2.77 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr9_+_137979506 | 2.76 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr11_+_46299199 | 2.75 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr12_+_6933660 | 2.75 |
ENST00000545321.1
|
GPR162
|
G protein-coupled receptor 162 |
| chr21_-_43916433 | 2.74 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr19_+_30017406 | 2.71 |
ENST00000335523.7
|
VSTM2B
|
V-set and transmembrane domain containing 2B |
| chr16_-_31021717 | 2.69 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr3_-_160116995 | 2.68 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr10_-_61469837 | 2.68 |
ENST00000395348.3
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr21_-_43916296 | 2.67 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr15_-_74045088 | 2.67 |
ENST00000569673.1
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr11_-_12030629 | 2.61 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr12_+_130822606 | 2.61 |
ENST00000546060.1
ENST00000539400.1 |
PIWIL1
|
piwi-like RNA-mediated gene silencing 1 |
| chr3_-_160117035 | 2.59 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr3_-_160117301 | 2.55 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chrX_-_92928557 | 2.54 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr3_+_112051994 | 2.53 |
ENST00000473539.1
ENST00000315711.8 ENST00000383681.3 |
CD200
|
CD200 molecule |
| chr20_-_30458019 | 2.51 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr5_+_169931249 | 2.51 |
ENST00000520740.1
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr3_+_63428982 | 2.49 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr7_+_75831181 | 2.48 |
ENST00000388802.4
ENST00000326382.8 |
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr11_-_12030681 | 2.48 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr14_+_33408449 | 2.48 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr15_+_43886057 | 2.47 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_-_83378638 | 2.47 |
ENST00000261722.3
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr15_+_43985725 | 2.47 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_+_1795012 | 2.46 |
ENST00000481110.2
ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr7_-_107880508 | 2.46 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_-_12031273 | 2.45 |
ENST00000525493.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_+_61520075 | 2.41 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr20_-_30458432 | 2.40 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr16_+_66461175 | 2.40 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr4_-_139163491 | 2.38 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr13_+_88324870 | 2.38 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr22_+_31518938 | 2.38 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr11_+_119019722 | 2.36 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr1_-_40782938 | 2.36 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr19_-_49149553 | 2.34 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr3_-_133614297 | 2.33 |
ENST00000486858.1
ENST00000477759.1 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr10_+_116853201 | 2.33 |
ENST00000527407.1
|
ATRNL1
|
attractin-like 1 |
| chr12_+_130822417 | 2.32 |
ENST00000245255.3
|
PIWIL1
|
piwi-like RNA-mediated gene silencing 1 |
| chr14_+_24540761 | 2.29 |
ENST00000559207.1
|
CPNE6
|
copine VI (neuronal) |
| chr4_+_41258786 | 2.29 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr11_-_12030130 | 2.26 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr3_-_62861012 | 2.26 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr9_+_129987488 | 2.24 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr20_-_30458491 | 2.24 |
ENST00000339738.5
|
DUSP15
|
dual specificity phosphatase 15 |
| chr12_-_21487829 | 2.21 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr9_-_120177342 | 2.21 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr22_+_26565440 | 2.18 |
ENST00000404234.3
ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L
|
seizure related 6 homolog (mouse)-like |
| chr16_+_56225248 | 2.16 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr22_-_38851205 | 2.15 |
ENST00000303592.3
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr8_-_40755333 | 2.15 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr19_-_36001286 | 2.15 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr14_-_103987679 | 2.14 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr6_+_30850697 | 2.14 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_223537401 | 2.14 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr12_-_120189900 | 2.14 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr19_-_49945617 | 2.12 |
ENST00000600601.1
ENST00000543531.1 |
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr17_+_11144580 | 2.11 |
ENST00000441885.3
ENST00000432116.3 ENST00000409168.3 |
SHISA6
|
shisa family member 6 |
| chr3_+_63428752 | 2.11 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr1_+_181452678 | 2.10 |
ENST00000367570.1
ENST00000526775.1 ENST00000357570.5 ENST00000358338.5 ENST00000367567.4 |
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr3_-_47619623 | 2.10 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr12_+_50451331 | 2.07 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr3_-_133614467 | 2.05 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr2_+_27371866 | 2.04 |
ENST00000296096.5
|
TCF23
|
transcription factor 23 |
| chr17_+_48610074 | 2.03 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr17_+_48609903 | 2.03 |
ENST00000268933.3
|
EPN3
|
epsin 3 |
| chr16_-_30023615 | 2.01 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr3_+_6902794 | 2.01 |
ENST00000357716.4
ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7
|
glutamate receptor, metabotropic 7 |
| chr19_-_55677920 | 2.00 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr6_+_149068464 | 1.99 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr4_-_7044657 | 1.98 |
ENST00000310085.4
|
CCDC96
|
coiled-coil domain containing 96 |
| chr6_-_31846744 | 1.98 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr4_-_66535653 | 1.97 |
ENST00000354839.4
ENST00000432638.2 |
EPHA5
|
EPH receptor A5 |
| chr5_+_92919043 | 1.97 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_+_43809797 | 1.96 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr19_-_36001386 | 1.96 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr2_+_220495800 | 1.95 |
ENST00000413743.1
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chrX_-_74742846 | 1.92 |
ENST00000373361.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr18_-_35145728 | 1.92 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr3_-_123123407 | 1.91 |
ENST00000466617.1
|
ADCY5
|
adenylate cyclase 5 |
| chr1_-_190443931 | 1.91 |
ENST00000445957.2
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr1_+_41827594 | 1.90 |
ENST00000372591.1
|
FOXO6
|
forkhead box O6 |
| chr2_+_210444748 | 1.90 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_+_1795508 | 1.90 |
ENST00000260795.2
ENST00000352904.1 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr4_+_30723003 | 1.89 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr5_+_137419581 | 1.87 |
ENST00000506684.1
ENST00000504809.1 ENST00000398754.1 |
WNT8A
|
wingless-type MMTV integration site family, member 8A |
| chr16_+_22825475 | 1.87 |
ENST00000261374.3
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chrX_+_12156582 | 1.87 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr15_-_45422056 | 1.87 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr15_+_71185148 | 1.87 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr9_+_71320557 | 1.86 |
ENST00000541509.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr7_-_4998802 | 1.84 |
ENST00000406755.1
ENST00000404774.3 ENST00000401401.3 |
MMD2
|
monocyte to macrophage differentiation-associated 2 |
| chr10_-_50122277 | 1.84 |
ENST00000374160.3
|
LRRC18
|
leucine rich repeat containing 18 |
| chr9_+_130478345 | 1.84 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr2_-_27603582 | 1.83 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chrX_+_153029633 | 1.83 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr11_-_64410787 | 1.83 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr1_+_15272271 | 1.83 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr2_+_120302041 | 1.83 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr3_-_123168551 | 1.81 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr1_-_72566613 | 1.80 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr18_-_35145689 | 1.80 |
ENST00000591287.1
ENST00000601019.1 ENST00000601392.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr6_-_150185156 | 1.79 |
ENST00000239367.2
ENST00000367368.2 |
LRP11
|
low density lipoprotein receptor-related protein 11 |
| chr17_-_41910505 | 1.79 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr9_-_117420052 | 1.78 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr6_+_43211418 | 1.78 |
ENST00000259750.4
|
TTBK1
|
tau tubulin kinase 1 |
| chr19_+_4304685 | 1.78 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr6_-_34073743 | 1.76 |
ENST00000609222.1
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr15_+_45422178 | 1.75 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr19_+_51153045 | 1.75 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr19_-_55677999 | 1.75 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr11_+_111411384 | 1.74 |
ENST00000375615.3
ENST00000525126.1 ENST00000436913.2 ENST00000533265.1 |
LAYN
|
layilin |
| chr19_+_48867652 | 1.74 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr5_+_149546334 | 1.73 |
ENST00000231656.8
|
CDX1
|
caudal type homeobox 1 |
| chr17_+_48610042 | 1.73 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr9_-_72287191 | 1.72 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr14_+_100204027 | 1.72 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr16_-_31021921 | 1.72 |
ENST00000215095.5
|
STX1B
|
syntaxin 1B |
| chr7_+_94537542 | 1.71 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr20_+_30458431 | 1.70 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr4_-_66536057 | 1.69 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr9_+_71320596 | 1.69 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr20_-_62129163 | 1.68 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr9_-_94712434 | 1.68 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr5_+_140220769 | 1.68 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr7_+_139208413 | 1.67 |
ENST00000422142.2
|
CLEC2L
|
C-type lectin domain family 2, member L |
| chr16_+_1756162 | 1.67 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr10_+_127585093 | 1.67 |
ENST00000368695.1
ENST00000368693.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr17_+_48638371 | 1.66 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr1_+_154540246 | 1.66 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr6_-_165723088 | 1.65 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr1_-_204654826 | 1.65 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr10_-_21463116 | 1.65 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr15_+_35838396 | 1.65 |
ENST00000501169.2
|
DPH6-AS1
|
DPH6 antisense RNA 1 (head to head) |
| chr1_-_233431458 | 1.64 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr3_-_133614597 | 1.64 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr7_-_642261 | 1.63 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr6_-_46459675 | 1.63 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr1_-_103574024 | 1.63 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr4_+_62066941 | 1.62 |
ENST00000512091.2
|
LPHN3
|
latrophilin 3 |
| chr19_-_55953704 | 1.62 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr1_-_98510843 | 1.62 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr7_+_73082152 | 1.61 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr17_+_40118805 | 1.61 |
ENST00000591072.1
ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr20_-_30458354 | 1.61 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr6_-_49755019 | 1.60 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr2_+_120301997 | 1.59 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr19_-_51529849 | 1.58 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chrX_-_55515635 | 1.58 |
ENST00000500968.3
|
USP51
|
ubiquitin specific peptidase 51 |
| chr6_-_114664180 | 1.57 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr4_+_657485 | 1.57 |
ENST00000471824.2
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 16.7 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 1.2 | 15.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.1 | 3.2 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 1.0 | 6.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.0 | 4.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.0 | 2.9 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.9 | 0.9 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.8 | 7.4 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.8 | 5.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.8 | 2.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 2.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.7 | 2.7 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.7 | 2.0 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.6 | 1.9 | GO:0044335 | neural crest cell fate commitment(GO:0014034) canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.6 | 2.3 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.6 | 3.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.5 | 3.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.5 | 3.6 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.5 | 1.5 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 4.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 2.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.4 | 2.7 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 4.3 | GO:0015820 | leucine transport(GO:0015820) |
| 0.4 | 5.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.4 | 1.6 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 4.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 0.7 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.3 | 1.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.3 | 8.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 0.7 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.3 | 5.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 1.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.3 | 2.5 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.3 | 1.9 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.3 | 1.9 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 1.2 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.3 | 4.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 1.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.3 | 1.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.3 | 6.7 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.3 | 0.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.3 | 0.3 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 1.7 | GO:0030047 | actin modification(GO:0030047) |
| 0.3 | 7.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 2.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 2.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 3.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 1.9 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.3 | 1.1 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.3 | 1.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 1.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 4.8 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 5.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 | 11.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 1.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.3 | 2.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.3 | 2.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.3 | 1.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 3.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 1.7 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 0.7 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.2 | 1.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 1.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 3.8 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 1.5 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 3.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 1.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.6 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 3.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 3.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 11.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.8 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.2 | 2.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 14.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.9 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 1.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 10.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.2 | 8.8 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.2 | 1.2 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.7 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 1.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 1.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 1.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.8 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.5 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.2 | 0.5 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 3.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 0.9 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 2.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.2 | GO:0021555 | rhombomere development(GO:0021546) midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 4.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 1.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.4 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 2.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 2.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0061626 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) BMP signaling pathway involved in heart development(GO:0061312) pharyngeal arch artery morphogenesis(GO:0061626) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 1.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.7 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 7.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.7 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.6 | GO:2001076 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.4 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.2 | GO:1901258 | cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 2.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 2.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.7 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 3.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.3 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 1.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 1.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 3.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 1.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.4 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.1 | 0.3 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.6 | GO:0002690 | positive regulation of leukocyte chemotaxis(GO:0002690) |
| 0.1 | 1.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.2 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.1 | 1.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 2.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.9 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.1 | 2.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 6.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 6.5 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 5.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 0.4 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 1.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.7 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 1.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 1.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.0 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.8 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.9 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.4 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 2.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 3.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 | 0.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 3.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 2.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 1.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 2.1 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 2.2 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 3.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.8 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 1.9 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 4.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 4.9 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.5 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0035407 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.8 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 2.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 4.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 2.8 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.7 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 1.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.2 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 3.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 3.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 1.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.9 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.4 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.4 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 2.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.4 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0061371 | determination of heart left/right asymmetry(GO:0061371) |
| 0.0 | 1.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0003157 | endocardium development(GO:0003157) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.3 | GO:0031571 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.7 | 4.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 2.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 2.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 5.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 0.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 0.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 4.9 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 9.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 6.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 2.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 7.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 1.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 1.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 1.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 1.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 0.9 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.2 | 1.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.3 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 0.8 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.6 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.4 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 3.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 14.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 11.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 3.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 3.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 12.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 4.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 19.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 3.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 6.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 7.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.9 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 4.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 7.5 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 7.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 5.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 10.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 5.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 35.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 3.5 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:1990175 | EH domain binding(GO:1990175) |
| 1.0 | 6.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.0 | 4.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.9 | 3.6 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.8 | 2.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 1.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.7 | 1.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.7 | 7.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.6 | 3.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.6 | 2.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 2.3 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.6 | 5.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.6 | 5.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 1.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 4.8 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.5 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.5 | 14.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 5.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.5 | 2.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.5 | 1.9 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.4 | 1.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.4 | 1.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.2 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.4 | 1.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 3.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 1.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.4 | 1.9 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.4 | 2.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 1.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 2.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 3.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 2.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 3.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 3.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 1.7 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.3 | 1.8 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 6.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 8.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 2.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 2.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 2.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 4.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.7 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.2 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.2 | 4.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 3.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 2.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 2.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 1.2 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 1.4 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 0.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 1.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 3.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.9 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 11.4 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 2.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 12.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 3.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 3.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 3.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 6.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.7 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 7.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.3 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 2.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 2.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 2.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.4 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.2 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.1 | 1.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 7.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.5 | GO:0004114 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.6 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 8.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 4.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.4 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 5.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 4.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 4.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 3.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 4.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 5.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 11.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 4.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 6.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 14.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |