Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for ZBTB7A_ZBTB7C
Z-value: 1.17
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
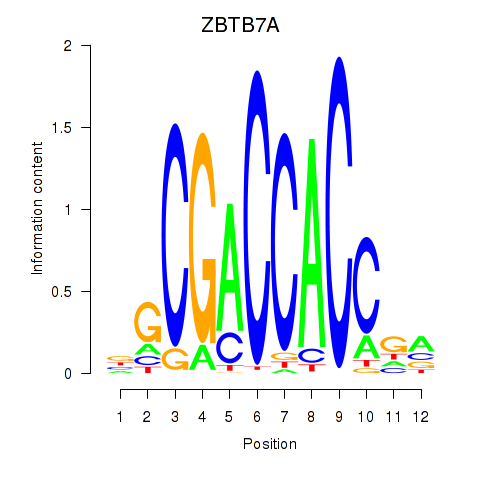
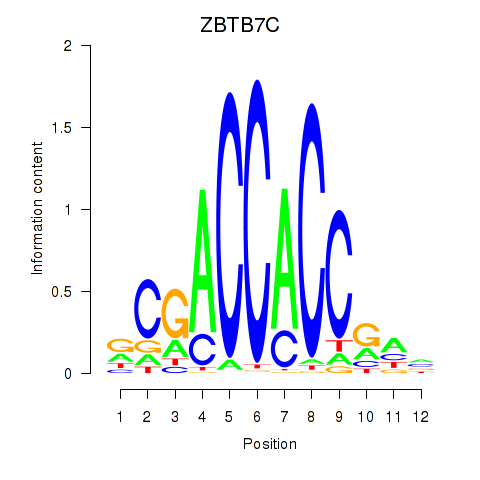
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4066890_4066943 | 0.16 | 3.9e-01 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45935663_45935793 | 0.15 | 4.2e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_57943781 | 3.83 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr14_+_29236269 | 2.45 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr5_-_132113063 | 2.28 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr5_-_132112921 | 2.03 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr5_-_132112907 | 2.01 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr5_-_132113690 | 1.66 |
ENST00000414594.1
|
SEPT8
|
septin 8 |
| chr5_-_114515734 | 1.61 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr20_+_36149602 | 1.61 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr2_+_198669365 | 1.55 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr5_-_132113036 | 1.53 |
ENST00000378706.1
|
SEPT8
|
septin 8 |
| chr4_-_16085340 | 1.39 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr4_-_16085314 | 1.38 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr19_+_35634146 | 1.36 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr13_+_96204961 | 1.34 |
ENST00000299339.2
|
CLDN10
|
claudin 10 |
| chr5_-_132113559 | 1.24 |
ENST00000448933.1
|
SEPT8
|
septin 8 |
| chr4_+_41258786 | 1.21 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr5_-_132113083 | 1.18 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr17_+_79989937 | 1.17 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr9_+_135037334 | 1.16 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr9_-_123476612 | 1.14 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr11_+_66045634 | 1.10 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr9_+_74764340 | 1.09 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr14_+_100150622 | 1.09 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr17_+_79990058 | 1.08 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_+_205538105 | 1.06 |
ENST00000367147.4
ENST00000539267.1 |
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr9_+_74764278 | 1.04 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr12_+_51985001 | 1.03 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr4_-_16085657 | 1.00 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr5_+_76506706 | 0.95 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr7_-_127672146 | 0.93 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr16_+_84002234 | 0.86 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr8_+_75896849 | 0.84 |
ENST00000520277.1
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chrX_-_20134713 | 0.83 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2 |
| chr4_-_185458660 | 0.83 |
ENST00000605270.1
|
RP11-326I11.4
|
RP11-326I11.4 |
| chr6_+_73331520 | 0.81 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr14_+_29234870 | 0.81 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr4_+_72053017 | 0.79 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_+_205538165 | 0.79 |
ENST00000536357.1
|
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr17_-_39769005 | 0.78 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr2_+_56411131 | 0.76 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr4_+_72052964 | 0.75 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_-_95007193 | 0.75 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr17_+_40834580 | 0.74 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chrX_+_152783131 | 0.73 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr13_-_30880979 | 0.73 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr5_-_87980753 | 0.73 |
ENST00000511014.2
|
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr1_+_169075554 | 0.72 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr12_+_13197218 | 0.72 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr12_+_51984657 | 0.71 |
ENST00000550891.1
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr8_-_144242020 | 0.71 |
ENST00000414417.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr7_+_30174574 | 0.71 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr20_-_23402028 | 0.71 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr5_-_138210977 | 0.70 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr12_+_50355647 | 0.70 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr11_-_12030681 | 0.68 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr8_-_144241432 | 0.68 |
ENST00000430474.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr8_-_144241664 | 0.68 |
ENST00000342752.4
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr17_-_42441204 | 0.68 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr15_+_27112948 | 0.68 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr17_+_64961026 | 0.68 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr3_-_179754556 | 0.63 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr14_+_33408449 | 0.63 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr5_-_2751762 | 0.62 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr22_+_21369316 | 0.62 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr17_+_43972010 | 0.61 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr8_+_75896731 | 0.61 |
ENST00000262207.4
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr1_+_38273818 | 0.60 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr17_-_39674668 | 0.60 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr14_-_22005062 | 0.59 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr5_-_138211051 | 0.59 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_43152191 | 0.59 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_-_235405168 | 0.58 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chrX_-_51812268 | 0.58 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr14_-_22005018 | 0.58 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_38273988 | 0.57 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr9_-_133814455 | 0.56 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr5_+_2752216 | 0.55 |
ENST00000457752.2
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr4_-_83719983 | 0.55 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr6_-_30043539 | 0.53 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr11_-_62752429 | 0.52 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_62752455 | 0.52 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr19_+_38755237 | 0.51 |
ENST00000587516.1
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_+_45971246 | 0.51 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr8_-_9762871 | 0.49 |
ENST00000521242.1
|
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr22_-_38480100 | 0.48 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr14_-_54955376 | 0.48 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr8_+_73449625 | 0.48 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr19_-_48673465 | 0.47 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr12_-_120805872 | 0.47 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr19_+_38755042 | 0.47 |
ENST00000301244.7
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr20_-_58508702 | 0.46 |
ENST00000357552.3
ENST00000425931.1 |
SYCP2
|
synaptonemal complex protein 2 |
| chr19_-_12945362 | 0.46 |
ENST00000590404.1
ENST00000592204.1 |
RTBDN
|
retbindin |
| chr9_+_132934835 | 0.46 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr14_-_54955721 | 0.45 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr11_-_62752162 | 0.45 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr19_+_38755203 | 0.44 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr3_-_33759541 | 0.43 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_+_4714792 | 0.42 |
ENST00000378190.3
|
AJAP1
|
adherens junctions associated protein 1 |
| chr19_-_56988677 | 0.42 |
ENST00000504904.3
ENST00000292069.6 |
ZNF667
|
zinc finger protein 667 |
| chr14_-_102771516 | 0.42 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr19_+_49622646 | 0.41 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr3_+_108308513 | 0.39 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr20_-_58508359 | 0.39 |
ENST00000446834.1
|
SYCP2
|
synaptonemal complex protein 2 |
| chrX_-_119603138 | 0.38 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr16_-_27895891 | 0.38 |
ENST00000569166.1
|
GSG1L
|
GSG1-like |
| chr5_-_33984741 | 0.37 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr3_+_108308559 | 0.37 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr7_-_127671674 | 0.37 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr3_-_33759699 | 0.36 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_-_64751227 | 0.35 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr12_+_132312931 | 0.35 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr15_-_64126084 | 0.34 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr15_+_76352178 | 0.34 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr19_+_40697514 | 0.33 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr5_+_102455968 | 0.33 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_-_122792919 | 0.33 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr11_-_1330834 | 0.33 |
ENST00000525159.1
ENST00000317204.6 ENST00000542915.1 ENST00000527938.1 ENST00000530541.1 ENST00000263646.7 |
TOLLIP
|
toll interacting protein |
| chr14_+_70233810 | 0.33 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr8_+_26240666 | 0.33 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_-_102771462 | 0.32 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr6_-_34113856 | 0.32 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr6_+_31496494 | 0.32 |
ENST00000376191.2
|
MCCD1
|
mitochondrial coiled-coil domain 1 |
| chr4_-_83719884 | 0.32 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr8_-_30891078 | 0.31 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr17_-_8534067 | 0.31 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr17_-_8534031 | 0.30 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr11_-_66115032 | 0.27 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr16_+_2521500 | 0.27 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr8_-_30890710 | 0.27 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr15_-_23932437 | 0.27 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr7_-_51384451 | 0.27 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr22_-_46373004 | 0.26 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr17_-_40835076 | 0.26 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr14_-_22005197 | 0.26 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_+_61600682 | 0.26 |
ENST00000581784.1
ENST00000456941.2 ENST00000580652.1 ENST00000314672.5 ENST00000583023.1 |
KCNH6
|
potassium voltage-gated channel, subfamily H (eag-related), member 6 |
| chrX_+_17393543 | 0.24 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr17_+_29158962 | 0.23 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr2_+_37311588 | 0.23 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr3_-_15469006 | 0.23 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr2_+_25015968 | 0.23 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr6_-_166755995 | 0.22 |
ENST00000361731.3
|
SFT2D1
|
SFT2 domain containing 1 |
| chr2_+_37311645 | 0.22 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr11_+_48002279 | 0.22 |
ENST00000534219.1
ENST00000527952.1 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr6_-_46459675 | 0.22 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr1_-_38273840 | 0.21 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr2_+_242255297 | 0.21 |
ENST00000401990.1
ENST00000407971.1 ENST00000436795.1 ENST00000411484.1 ENST00000434955.1 ENST00000402092.2 ENST00000441533.1 ENST00000443492.1 ENST00000437066.1 ENST00000429791.1 |
SEPT2
|
septin 2 |
| chr10_+_49514698 | 0.21 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr14_-_58332774 | 0.21 |
ENST00000556826.1
|
SLC35F4
|
solute carrier family 35, member F4 |
| chr14_-_71276211 | 0.21 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_+_47630255 | 0.21 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr8_-_101963482 | 0.21 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_+_179322573 | 0.20 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr15_-_83876758 | 0.20 |
ENST00000299633.4
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr2_+_242255275 | 0.20 |
ENST00000391971.2
|
SEPT2
|
septin 2 |
| chr11_+_48002076 | 0.20 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr19_-_30199516 | 0.20 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr2_-_46769694 | 0.20 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr1_+_162467595 | 0.19 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr1_+_110754094 | 0.19 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chrX_-_153141783 | 0.19 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr15_-_33486897 | 0.19 |
ENST00000320930.7
|
FMN1
|
formin 1 |
| chr14_+_59655369 | 0.19 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr20_-_35492048 | 0.19 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr11_-_117747327 | 0.19 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr20_+_44034804 | 0.19 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_+_25016282 | 0.18 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr8_+_26240414 | 0.18 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_-_101964231 | 0.17 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_164914640 | 0.17 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr17_-_58469329 | 0.17 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr10_+_14880364 | 0.17 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr18_+_74207477 | 0.17 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr4_-_119274121 | 0.17 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr5_-_443239 | 0.17 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr21_-_45079341 | 0.17 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr16_-_19896220 | 0.17 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr8_+_9413410 | 0.16 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr16_-_49315731 | 0.16 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr3_-_100120223 | 0.16 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr2_+_58655520 | 0.16 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr1_+_39571026 | 0.16 |
ENST00000524432.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr8_-_101963677 | 0.16 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chrX_+_133507327 | 0.16 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr3_-_15469045 | 0.15 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr16_+_89724188 | 0.15 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr12_-_49351148 | 0.15 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr1_+_19991780 | 0.15 |
ENST00000289753.1
|
HTR6
|
5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled |
| chr2_+_136289030 | 0.15 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr4_-_105416039 | 0.15 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chrX_+_133507283 | 0.15 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chr2_+_47630108 | 0.15 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr8_+_145202939 | 0.15 |
ENST00000423230.2
ENST00000398656.4 |
MROH1
|
maestro heat-like repeat family member 1 |
| chr20_+_37590942 | 0.15 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr20_-_61493115 | 0.15 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr4_-_5021164 | 0.15 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr2_+_48667983 | 0.15 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr8_-_74884511 | 0.15 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr19_-_5567996 | 0.15 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chrX_+_135579238 | 0.14 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr8_-_140715294 | 0.14 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.8 | 3.8 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.7 | 2.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.5 | 1.5 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.4 | 1.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 0.9 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 1.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.3 | 0.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 0.9 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 1.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.5 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.2 | 0.8 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 0.6 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 2.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.6 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.4 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.6 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 1.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.7 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.7 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.3 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 1.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 3.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 1.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.7 | GO:0060384 | gamma-aminobutyric acid signaling pathway(GO:0007214) innervation(GO:0060384) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) mitochondrial pyruvate transport(GO:0006850) pyruvate transmembrane transport(GO:1901475) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 3.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.4 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.3 | 1.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 0.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.4 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 3.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 1.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 1.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.2 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 3.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 14.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |