Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 1.76
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
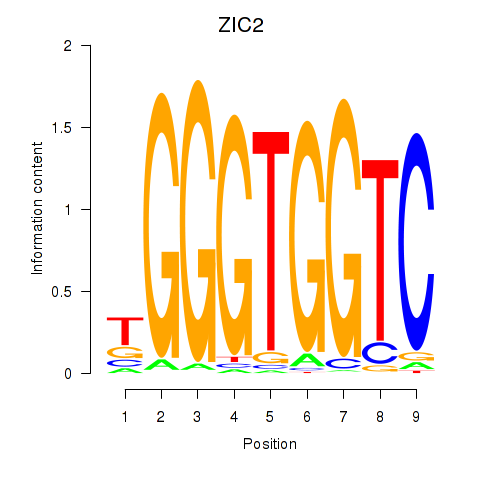
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
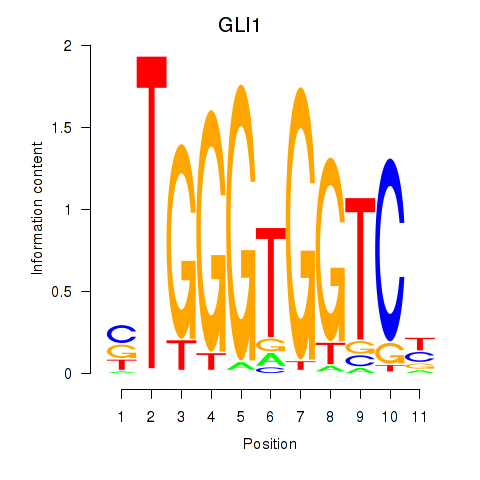
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57854274_57854274 | 0.29 | 1.0e-01 | Click! |
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | 0.23 | 2.1e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_111783919 | 8.37 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr1_+_47264711 | 7.89 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr11_-_111783595 | 7.46 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr11_+_111783450 | 6.86 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr5_-_168727713 | 6.62 |
ENST00000404867.3
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr11_+_111782934 | 6.58 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr5_-_168727786 | 6.30 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr5_-_44388899 | 6.29 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chr20_+_36149602 | 5.94 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr3_-_73673991 | 4.89 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chrX_+_90689810 | 4.68 |
ENST00000312600.3
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5 |
| chr1_-_40105617 | 4.30 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr8_-_17555164 | 4.13 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr20_+_43343476 | 3.60 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_+_153839149 | 3.52 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr9_-_124989804 | 3.44 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr10_-_95360983 | 3.40 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr20_+_43343886 | 3.31 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_+_137523086 | 3.26 |
ENST00000409968.1
|
THSD7B
|
thrombospondin, type I, domain containing 7B |
| chr21_-_27945562 | 3.22 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr6_-_26027480 | 3.22 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr17_-_74533734 | 3.21 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr14_+_101292445 | 3.20 |
ENST00000429159.2
ENST00000520714.1 ENST00000522771.2 ENST00000424076.3 ENST00000423456.1 ENST00000521404.1 ENST00000556736.1 ENST00000451743.2 ENST00000398518.2 ENST00000554639.1 ENST00000452120.2 ENST00000519709.1 ENST00000412736.2 |
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_-_45308616 | 3.17 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr7_+_119913688 | 3.08 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr5_+_71403061 | 3.05 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr4_-_5021164 | 3.05 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr1_-_171621815 | 3.03 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr19_+_676385 | 3.00 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr17_+_6926339 | 2.93 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr11_+_118478313 | 2.92 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr8_+_75736761 | 2.86 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr8_+_30244580 | 2.70 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr15_-_93616892 | 2.62 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr20_+_43343517 | 2.54 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_-_141704566 | 2.48 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr5_+_71403280 | 2.47 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr11_+_76493294 | 2.39 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr11_-_111784005 | 2.37 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B |
| chr21_-_28215332 | 2.35 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr16_+_86612112 | 2.35 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr19_-_35625765 | 2.35 |
ENST00000591633.1
|
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr2_+_217498105 | 2.31 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr6_+_163148973 | 2.26 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr11_-_12030681 | 2.17 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr5_+_52776449 | 2.14 |
ENST00000396947.3
|
FST
|
follistatin |
| chr17_+_6926381 | 2.14 |
ENST00000576705.1
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr9_+_109625378 | 2.13 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr17_-_74533963 | 2.07 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr11_-_111782696 | 2.04 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr4_-_186696425 | 2.03 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_116312411 | 2.02 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr11_-_75379612 | 2.02 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr4_-_187517928 | 1.99 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr11_-_111782484 | 1.99 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr6_+_43739697 | 1.98 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr5_+_52776228 | 1.98 |
ENST00000256759.3
|
FST
|
follistatin |
| chr19_+_45312347 | 1.97 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr9_-_34397800 | 1.94 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr11_-_11643540 | 1.92 |
ENST00000227756.4
|
GALNT18
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr11_-_12030130 | 1.81 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr19_+_45312310 | 1.78 |
ENST00000589651.1
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr6_-_29600559 | 1.76 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_130131907 | 1.70 |
ENST00000223215.4
ENST00000437945.1 |
MEST
|
mesoderm specific transcript |
| chr19_-_51456321 | 1.69 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr11_-_12030629 | 1.68 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr6_+_17281802 | 1.67 |
ENST00000509686.1
|
RBM24
|
RNA binding motif protein 24 |
| chr17_+_7608511 | 1.65 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr11_-_75379479 | 1.64 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr17_+_40834580 | 1.63 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr17_-_34122596 | 1.61 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr9_+_131174024 | 1.59 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr16_+_28875268 | 1.57 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr21_-_27945464 | 1.57 |
ENST00000400043.3
|
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr7_-_151107106 | 1.54 |
ENST00000334493.6
|
WDR86
|
WD repeat domain 86 |
| chr4_-_186696561 | 1.53 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_76422409 | 1.53 |
ENST00000600087.1
|
AC061992.1
|
Uncharacterized protein |
| chr3_+_159481464 | 1.51 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr21_-_28217721 | 1.51 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_-_64527425 | 1.49 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr4_-_186697044 | 1.49 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_-_30396802 | 1.48 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr1_+_164528866 | 1.46 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr19_-_51456344 | 1.46 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr10_+_24755416 | 1.44 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr1_-_183387723 | 1.44 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr19_-_41196534 | 1.41 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr20_+_62327996 | 1.41 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr19_-_40919271 | 1.40 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr19_-_41196458 | 1.38 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr1_-_182573514 | 1.37 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr16_+_86600857 | 1.37 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr10_-_90712520 | 1.37 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr14_-_23834411 | 1.35 |
ENST00000429593.2
|
EFS
|
embryonal Fyn-associated substrate |
| chr1_+_226736446 | 1.33 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr3_+_159481791 | 1.31 |
ENST00000460298.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_-_148758839 | 1.30 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr9_+_135285579 | 1.28 |
ENST00000343036.2
ENST00000393216.2 |
C9orf171
|
chromosome 9 open reading frame 171 |
| chr11_-_65640071 | 1.25 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr2_-_133427767 | 1.25 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr5_-_137878887 | 1.23 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr2_-_213403565 | 1.22 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr4_+_62067860 | 1.21 |
ENST00000514591.1
|
LPHN3
|
latrophilin 3 |
| chr11_-_119991589 | 1.20 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr6_+_125540951 | 1.20 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr3_+_170136642 | 1.18 |
ENST00000064724.3
ENST00000486975.1 |
CLDN11
|
claudin 11 |
| chr17_+_17876127 | 1.18 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr5_-_137090028 | 1.18 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr18_+_21529811 | 1.17 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_159825143 | 1.15 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_+_26915584 | 1.15 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr2_-_193059634 | 1.15 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr4_-_186696636 | 1.14 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr13_+_44947941 | 1.14 |
ENST00000379179.3
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr3_+_159481988 | 1.13 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chrX_+_70364667 | 1.13 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr16_+_86229728 | 1.12 |
ENST00000601250.1
|
LINC01082
|
long intergenic non-protein coding RNA 1082 |
| chr4_-_186696515 | 1.09 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_-_61124977 | 1.09 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr6_-_29600832 | 1.09 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr22_-_28197486 | 1.08 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr4_+_134070439 | 1.06 |
ENST00000264360.5
|
PCDH10
|
protocadherin 10 |
| chr1_-_38512450 | 1.06 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr11_-_30608413 | 1.04 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr11_+_45168182 | 1.04 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr7_-_108096765 | 0.98 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr2_+_25016282 | 0.96 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr13_-_30880979 | 0.96 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr3_+_113616317 | 0.95 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr7_-_108096822 | 0.95 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_205782304 | 0.95 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr10_-_97050777 | 0.94 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr6_-_19804973 | 0.94 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chrX_-_138287168 | 0.93 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr10_-_35104185 | 0.90 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr16_-_21452040 | 0.88 |
ENST00000521589.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr10_-_69455873 | 0.88 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr2_+_25015968 | 0.87 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr17_-_7137582 | 0.87 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr17_-_7137857 | 0.86 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr9_-_98079154 | 0.85 |
ENST00000433829.1
|
FANCC
|
Fanconi anemia, complementation group C |
| chr15_-_23891175 | 0.83 |
ENST00000532292.1
|
MAGEL2
|
MAGE-like 2 |
| chr14_-_51561784 | 0.82 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chrX_-_140271249 | 0.81 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr2_-_51259641 | 0.78 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr9_+_136399929 | 0.78 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr1_+_42846443 | 0.77 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr20_+_32782375 | 0.76 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr19_-_42758040 | 0.76 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr2_-_28113965 | 0.76 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr16_-_85146040 | 0.75 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr6_-_110500826 | 0.75 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr17_-_15496722 | 0.73 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr6_-_55739542 | 0.73 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr1_+_183155373 | 0.72 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr7_-_33842742 | 0.71 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr3_+_110790867 | 0.70 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr8_-_41522719 | 0.67 |
ENST00000335651.6
|
ANK1
|
ankyrin 1, erythrocytic |
| chr2_-_51259528 | 0.67 |
ENST00000404971.1
|
NRXN1
|
neurexin 1 |
| chr9_-_130639997 | 0.65 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr9_+_135285430 | 0.65 |
ENST00000393215.3
|
C9orf171
|
chromosome 9 open reading frame 171 |
| chrX_-_107979616 | 0.65 |
ENST00000372129.2
|
IRS4
|
insulin receptor substrate 4 |
| chr10_-_77161004 | 0.65 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr7_-_108097144 | 0.64 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr2_-_213403253 | 0.64 |
ENST00000260943.6
ENST00000402597.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr4_-_134070250 | 0.64 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr17_-_15244894 | 0.64 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr15_-_77712429 | 0.63 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_+_137717571 | 0.63 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr9_-_33264557 | 0.63 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr11_+_59224411 | 0.63 |
ENST00000300127.2
|
OR4D6
|
olfactory receptor, family 4, subfamily D, member 6 |
| chr6_+_43968306 | 0.63 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr1_-_217250231 | 0.62 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr14_+_96968802 | 0.61 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr3_+_6902794 | 0.60 |
ENST00000357716.4
ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7
|
glutamate receptor, metabotropic 7 |
| chr2_+_68694678 | 0.59 |
ENST00000303795.4
|
APLF
|
aprataxin and PNKP like factor |
| chr3_+_110790715 | 0.58 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chr1_+_32674675 | 0.56 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr10_-_77161650 | 0.56 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr10_-_77161533 | 0.54 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr9_+_34990219 | 0.54 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr22_+_38201114 | 0.54 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr17_-_17875688 | 0.54 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr3_+_110790590 | 0.54 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr2_-_51259292 | 0.53 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr16_+_57844549 | 0.53 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr11_-_65321198 | 0.53 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr12_+_65996599 | 0.53 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr7_-_4901625 | 0.52 |
ENST00000404991.1
|
PAPOLB
|
poly(A) polymerase beta (testis specific) |
| chr22_-_46373004 | 0.51 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr1_+_152943122 | 0.51 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr10_-_81203972 | 0.51 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr22_-_19165917 | 0.51 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr22_-_30783075 | 0.50 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr22_-_19166343 | 0.50 |
ENST00000215882.5
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr7_-_112726393 | 0.50 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chrX_+_51636629 | 0.49 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr10_-_18948156 | 0.49 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr19_-_30199516 | 0.49 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr8_-_41522779 | 0.47 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 12.9 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 2.1 | 6.3 | GO:0071335 | proximal/distal axis specification(GO:0009946) bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) secretion by lung epithelial cell involved in lung growth(GO:0061033) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.4 | 4.3 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.9 | 22.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.9 | 7.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.8 | 3.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.8 | 5.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.7 | 5.7 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.7 | 2.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.7 | 3.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.7 | 2.0 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.6 | 1.9 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.6 | 2.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.5 | 3.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 7.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 5.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.5 | 1.4 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.5 | 5.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.4 | 1.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 1.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.4 | 0.7 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.3 | 2.4 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.3 | 1.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.3 | 1.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 2.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 1.6 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 4.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 0.5 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.2 | 1.5 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 3.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 1.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 1.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 5.5 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 0.8 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.2 | 3.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 3.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.8 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 7.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 1.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 1.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 4.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.8 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.1 | 1.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 2.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 2.7 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.6 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 2.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 1.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.9 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.8 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 3.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 4.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.8 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.6 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 3.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.5 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.1 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 1.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.5 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.7 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 1.1 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 1.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 12.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.2 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 2.4 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.6 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 1.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.4 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 2.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 1.6 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.6 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.0 | GO:0019046 | release from viral latency(GO:0019046) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 22.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 3.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 3.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 3.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 3.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 4.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 3.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 4.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 4.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 49.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.3 | 5.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 1.3 | 12.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.3 | 3.8 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 1.1 | 4.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.7 | 22.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.6 | 3.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 2.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 1.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 2.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 3.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 8.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 7.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 1.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 11.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 1.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.2 | 3.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.8 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.2 | 1.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 7.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 3.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 3.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.8 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) collagen V binding(GO:0070052) |
| 0.0 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 3.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 4.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 3.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 6.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 6.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 19.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 3.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 14.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 7.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 5.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 4.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 2.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |