Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for ZKSCAN1
Z-value: 0.74
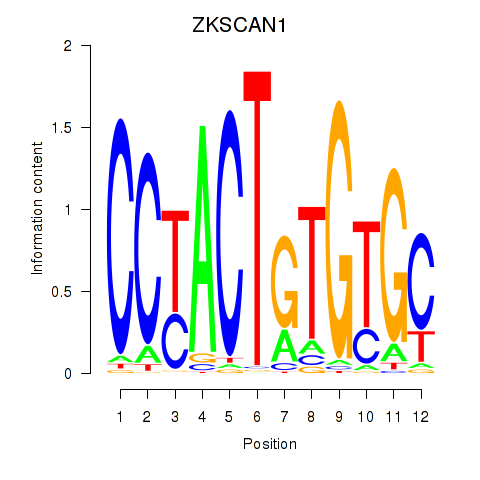
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | zinc finger with KRAB and SCAN domains 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN1 | hg19_v2_chr7_+_99613195_99613206 | -0.33 | 6.1e-02 | Click! |
Activity profile of ZKSCAN1 motif
Sorted Z-values of ZKSCAN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55660561 | 2.52 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr16_+_4364762 | 1.80 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr6_+_36922209 | 1.79 |
ENST00000373674.3
|
PI16
|
peptidase inhibitor 16 |
| chr11_-_73720122 | 1.76 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr10_-_76859247 | 1.68 |
ENST00000472493.2
ENST00000605915.1 ENST00000478873.2 |
DUSP13
|
dual specificity phosphatase 13 |
| chr16_-_11370330 | 1.55 |
ENST00000241808.4
ENST00000435245.2 |
PRM2
|
protamine 2 |
| chr11_-_73720276 | 1.30 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr20_+_43343476 | 1.28 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_+_127039075 | 1.22 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr3_-_195310802 | 1.17 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr5_+_174151536 | 1.12 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr20_+_43343517 | 1.09 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_-_50666431 | 1.08 |
ENST00000293405.3
|
IZUMO2
|
IZUMO family member 2 |
| chr20_+_44098346 | 1.08 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr19_+_6740888 | 1.07 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr16_-_46797149 | 1.03 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr1_+_16083098 | 0.93 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr18_+_71983048 | 0.92 |
ENST00000579455.1
|
C18orf63
|
chromosome 18 open reading frame 63 |
| chr1_+_16083123 | 0.92 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_16083154 | 0.90 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_-_15402498 | 0.90 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chrX_-_139047669 | 0.89 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr15_+_75494214 | 0.85 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr5_-_172756506 | 0.84 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr20_+_44098385 | 0.83 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr5_+_149980622 | 0.81 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr5_+_155753745 | 0.78 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr4_-_13549417 | 0.75 |
ENST00000501050.1
|
AC006445.8
|
long intergenic non-protein coding RNA 1096 |
| chr10_+_106113515 | 0.74 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr19_-_50666404 | 0.72 |
ENST00000600293.1
|
IZUMO2
|
IZUMO family member 2 |
| chr1_+_113009163 | 0.70 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr9_-_86153218 | 0.70 |
ENST00000304195.3
ENST00000376438.1 |
FRMD3
|
FERM domain containing 3 |
| chr11_-_22881972 | 0.69 |
ENST00000532798.2
|
CCDC179
|
coiled-coil domain containing 179 |
| chr20_-_56265680 | 0.68 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_16348366 | 0.66 |
ENST00000375692.1
ENST00000420078.1 |
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr2_+_74120094 | 0.65 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_+_72742178 | 0.63 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr7_+_72742162 | 0.63 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr10_-_46640660 | 0.63 |
ENST00000395727.2
ENST00000509900.1 ENST00000503851.1 ENST00000506080.1 ENST00000513266.1 ENST00000502705.1 ENST00000505814.1 ENST00000509774.1 ENST00000511769.1 ENST00000509599.1 ENST00000513156.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr15_+_78370140 | 0.62 |
ENST00000409568.2
|
SH2D7
|
SH2 domain containing 7 |
| chr12_+_9066472 | 0.57 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr1_+_16370271 | 0.56 |
ENST00000375679.4
|
CLCNKB
|
chloride channel, voltage-sensitive Kb |
| chr14_+_63671577 | 0.56 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr7_-_72742085 | 0.55 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr14_-_76447336 | 0.55 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr16_+_19729586 | 0.51 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr19_-_344786 | 0.51 |
ENST00000264819.4
|
MIER2
|
mesoderm induction early response 1, family member 2 |
| chr5_-_127873659 | 0.50 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr1_+_16348497 | 0.49 |
ENST00000439316.2
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr17_+_26800648 | 0.47 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_27709816 | 0.47 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr17_+_26800756 | 0.46 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_151804314 | 0.44 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr19_-_7764960 | 0.44 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr15_+_75498739 | 0.43 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr2_+_1488435 | 0.42 |
ENST00000446278.1
ENST00000469607.1 |
TPO
|
thyroid peroxidase |
| chr1_-_151804222 | 0.42 |
ENST00000392697.3
|
RORC
|
RAR-related orphan receptor C |
| chr17_+_16945820 | 0.42 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr14_+_38033252 | 0.39 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr8_+_22102626 | 0.38 |
ENST00000519237.1
ENST00000397802.4 |
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr22_+_33197683 | 0.38 |
ENST00000266085.6
|
TIMP3
|
TIMP metallopeptidase inhibitor 3 |
| chr1_-_205391178 | 0.37 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr15_-_49102781 | 0.35 |
ENST00000558591.1
|
CEP152
|
centrosomal protein 152kDa |
| chr1_-_27709793 | 0.35 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr14_+_101361107 | 0.35 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr6_+_35265586 | 0.35 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr19_-_3062465 | 0.35 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chrX_-_130423200 | 0.34 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr10_-_126432821 | 0.34 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr4_+_41540160 | 0.33 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_+_8736232 | 0.33 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr17_+_36858694 | 0.32 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr11_+_72975524 | 0.32 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_64739358 | 0.32 |
ENST00000301896.5
ENST00000530444.1 |
C11orf85
|
chromosome 11 open reading frame 85 |
| chr9_+_100069933 | 0.30 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr1_+_236958554 | 0.30 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr9_+_138413277 | 0.30 |
ENST00000263598.2
ENST00000371781.3 |
LCN1
|
lipocalin 1 |
| chr9_-_123639600 | 0.30 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr22_+_20861858 | 0.29 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr2_+_46926326 | 0.28 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr19_+_13135731 | 0.28 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_+_87563557 | 0.27 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr3_+_63898275 | 0.26 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr10_-_126432619 | 0.26 |
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B |
| chr11_-_64739542 | 0.26 |
ENST00000536065.1
|
C11orf85
|
chromosome 11 open reading frame 85 |
| chr12_-_123450986 | 0.26 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr19_+_16830815 | 0.25 |
ENST00000549814.1
|
NWD1
|
NACHT and WD repeat domain containing 1 |
| chr6_+_157099036 | 0.25 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr11_+_72975578 | 0.25 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr19_+_39647271 | 0.24 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr15_-_49103184 | 0.24 |
ENST00000399334.3
ENST00000325747.5 |
CEP152
|
centrosomal protein 152kDa |
| chr9_-_16253112 | 0.23 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr11_+_72975559 | 0.22 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr6_+_151815143 | 0.22 |
ENST00000239374.7
ENST00000367290.5 |
CCDC170
|
coiled-coil domain containing 170 |
| chr16_+_616995 | 0.22 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chr14_+_103851712 | 0.22 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr15_-_49103235 | 0.22 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr5_-_131562935 | 0.21 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chrX_+_109245863 | 0.21 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr12_+_113416340 | 0.21 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr19_+_13135790 | 0.20 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr15_+_78384901 | 0.20 |
ENST00000328828.5
|
SH2D7
|
SH2 domain containing 7 |
| chr1_+_170115142 | 0.20 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr21_+_46117087 | 0.19 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr19_+_39138320 | 0.19 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr1_+_203256898 | 0.19 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chr5_+_137774706 | 0.19 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr3_-_64673289 | 0.19 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr10_-_112678692 | 0.18 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chrX_-_130423386 | 0.18 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr1_+_32083301 | 0.18 |
ENST00000403528.2
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr1_+_62901968 | 0.18 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr17_+_18163848 | 0.17 |
ENST00000323019.4
ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2
|
mitochondrial elongation factor 2 |
| chr20_+_36932521 | 0.17 |
ENST00000262865.4
|
BPI
|
bactericidal/permeability-increasing protein |
| chr13_+_76445187 | 0.16 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr10_+_98592674 | 0.16 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr16_+_19125252 | 0.16 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr11_-_72070206 | 0.16 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr17_-_73150629 | 0.15 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr19_+_39138271 | 0.15 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr17_-_73150599 | 0.14 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr20_-_3644046 | 0.14 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr5_+_135394840 | 0.14 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_-_51796987 | 0.14 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr19_+_999601 | 0.14 |
ENST00000594393.1
|
AC004528.1
|
Uncharacterized protein |
| chr16_+_67562702 | 0.14 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr10_+_112679301 | 0.14 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr10_+_118350522 | 0.14 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr7_+_87563458 | 0.14 |
ENST00000398204.4
|
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr14_+_54976603 | 0.13 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr2_+_101179152 | 0.13 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr16_+_3550924 | 0.13 |
ENST00000576634.1
ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1
|
clusterin associated protein 1 |
| chr6_-_30640811 | 0.13 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_+_64808675 | 0.13 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr9_-_114090713 | 0.13 |
ENST00000302681.1
ENST00000374428.1 |
OR2K2
|
olfactory receptor, family 2, subfamily K, member 2 |
| chr12_-_112037306 | 0.12 |
ENST00000535949.1
ENST00000542287.2 ENST00000377617.3 ENST00000550104.1 |
ATXN2
|
ataxin 2 |
| chr1_+_156551157 | 0.12 |
ENST00000368237.3
|
TTC24
|
tetratricopeptide repeat domain 24 |
| chr7_-_72439997 | 0.12 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr6_+_10634158 | 0.12 |
ENST00000379591.3
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr7_+_75024903 | 0.12 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr19_-_3062881 | 0.11 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr2_-_37193606 | 0.11 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr6_-_30640761 | 0.11 |
ENST00000415603.1
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr3_-_71834318 | 0.11 |
ENST00000353065.3
|
PROK2
|
prokineticin 2 |
| chr10_-_112678904 | 0.10 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr4_-_186732048 | 0.10 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_-_19051993 | 0.09 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr6_+_10633993 | 0.09 |
ENST00000417671.1
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr4_+_41361616 | 0.08 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_45140400 | 0.07 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr14_-_67981870 | 0.07 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr15_+_40731920 | 0.07 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr10_+_118350468 | 0.06 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chrX_-_40036520 | 0.06 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr17_-_46716647 | 0.06 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr2_+_69201705 | 0.05 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr3_-_179322416 | 0.04 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr11_-_61197187 | 0.03 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr21_+_45993606 | 0.03 |
ENST00000400374.3
|
KRTAP10-4
|
keratin associated protein 10-4 |
| chr6_+_32944119 | 0.03 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr4_+_39460689 | 0.03 |
ENST00000381846.1
|
LIAS
|
lipoic acid synthetase |
| chr12_+_113416265 | 0.03 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr3_-_179322436 | 0.02 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr10_-_88281494 | 0.02 |
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila) |
| chr14_-_96743060 | 0.02 |
ENST00000554299.1
|
DKFZP434O1614
|
DKFZP434O1614 |
| chr11_+_46366918 | 0.02 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr19_-_3063099 | 0.01 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chrX_-_130423240 | 0.00 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.3 | 1.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 2.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.5 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.3 | 1.8 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.2 | 1.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 3.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 2.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 1.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.9 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 1.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0035411 | catenin import into nucleus(GO:0035411) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 0.9 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.3 | 1.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 2.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 2.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 1.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 3.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |