Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
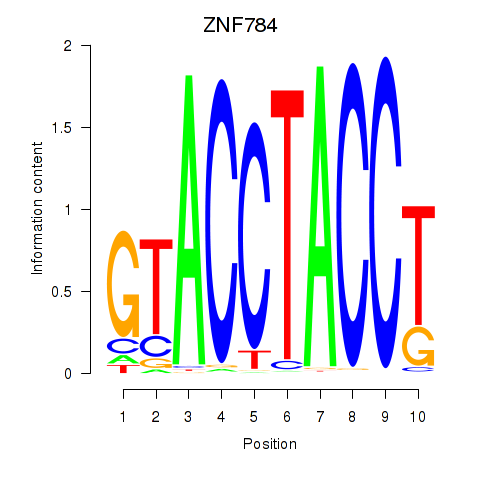
Results for ZNF784
Z-value: 1.37
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg19_v2_chr19_-_56135928_56135967 | -0.42 | 1.8e-02 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_42498369 | 5.67 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_-_42498231 | 5.34 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr3_-_42306248 | 3.61 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr8_+_80523321 | 3.40 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr4_-_6202247 | 2.69 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr3_-_47619623 | 2.58 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr4_+_158142750 | 2.47 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr1_+_205012293 | 2.38 |
ENST00000331830.4
|
CNTN2
|
contactin 2 (axonal) |
| chr5_+_161275320 | 2.31 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_+_50575292 | 2.19 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_+_50574585 | 2.17 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_+_161274940 | 2.16 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_-_6202291 | 2.09 |
ENST00000282924.5
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr4_-_90758227 | 2.01 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_-_232791038 | 1.96 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr3_+_183993797 | 1.93 |
ENST00000359140.4
ENST00000404464.3 ENST00000357474.5 |
ECE2
|
endothelin converting enzyme 2 |
| chr4_-_90758118 | 1.88 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_+_129338008 | 1.81 |
ENST00000442111.2
ENST00000281703.6 |
GLT1D1
|
glycosyltransferase 1 domain containing 1 |
| chr12_-_62585203 | 1.78 |
ENST00000551449.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr4_-_187476464 | 1.69 |
ENST00000509111.1
|
RP11-215A19.2
|
RP11-215A19.2 |
| chr19_+_7562431 | 1.62 |
ENST00000361664.2
|
C19orf45
|
chromosome 19 open reading frame 45 |
| chr4_+_158141899 | 1.62 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chrX_-_6146876 | 1.61 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr9_+_34957477 | 1.61 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chr4_+_158141806 | 1.58 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr22_+_21321531 | 1.58 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr8_-_139509065 | 1.56 |
ENST00000395297.1
|
FAM135B
|
family with sequence similarity 135, member B |
| chr3_+_85008089 | 1.55 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr4_-_90756769 | 1.55 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr7_+_142636603 | 1.55 |
ENST00000409607.3
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr11_+_73358690 | 1.54 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr22_-_44708731 | 1.49 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr18_+_905104 | 1.48 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr4_+_158141843 | 1.46 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr11_-_18765389 | 1.45 |
ENST00000477854.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr3_+_50712672 | 1.43 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr7_+_142636440 | 1.42 |
ENST00000458732.1
|
C7orf34
|
chromosome 7 open reading frame 34 |
| chr3_-_62860704 | 1.40 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr2_-_40739501 | 1.40 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr8_-_57358432 | 1.39 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr6_+_123100620 | 1.38 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr11_+_73358594 | 1.36 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr7_-_150652924 | 1.34 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr22_+_21321447 | 1.32 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr12_-_49525175 | 1.32 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chrX_+_144908928 | 1.31 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr10_+_18240834 | 1.31 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr18_-_31802056 | 1.30 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr18_-_31802282 | 1.28 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr5_-_174871136 | 1.28 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr11_-_5248294 | 1.28 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr7_+_26332645 | 1.27 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr15_+_40532058 | 1.25 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr7_+_8474693 | 1.25 |
ENST00000438764.1
|
NXPH1
|
neurexophilin 1 |
| chr15_+_40453204 | 1.25 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr11_-_12030130 | 1.23 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr7_+_8474817 | 1.19 |
ENST00000429542.1
|
NXPH1
|
neurexophilin 1 |
| chr12_-_13256571 | 1.18 |
ENST00000545401.1
ENST00000432710.2 ENST00000351606.6 |
GSG1
|
germ cell associated 1 |
| chr19_+_3880581 | 1.16 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr17_+_73717516 | 1.16 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr10_+_18240753 | 1.16 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr8_-_101117847 | 1.15 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chr10_+_18240814 | 1.15 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr8_-_101118210 | 1.12 |
ENST00000360863.6
|
RGS22
|
regulator of G-protein signaling 22 |
| chr20_+_20348740 | 1.12 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr15_+_40531621 | 1.09 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr4_-_90757364 | 1.09 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_73717551 | 1.09 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr11_-_134281812 | 1.06 |
ENST00000392580.1
ENST00000312527.4 |
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr8_-_101118185 | 1.06 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr17_+_73717407 | 1.04 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr1_+_173837214 | 1.04 |
ENST00000367704.1
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr8_+_85095497 | 1.04 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr3_-_39321512 | 1.03 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr12_-_13256593 | 1.03 |
ENST00000542415.1
ENST00000324458.8 |
GSG1
|
germ cell associated 1 |
| chr4_-_153274078 | 1.02 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_+_41221794 | 1.01 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr17_-_50237343 | 1.01 |
ENST00000575181.1
ENST00000570565.1 |
CA10
|
carbonic anhydrase X |
| chr12_-_58131931 | 1.00 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_-_161695042 | 0.98 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_-_85156090 | 0.96 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chrX_-_153095945 | 0.96 |
ENST00000164640.4
|
PDZD4
|
PDZ domain containing 4 |
| chr15_+_59664884 | 0.96 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chrX_-_153095813 | 0.94 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr14_-_55658323 | 0.93 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr5_+_125706998 | 0.93 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr2_+_14772810 | 0.93 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr12_-_12837423 | 0.93 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr6_+_123100853 | 0.92 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr1_-_85155939 | 0.92 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_186649543 | 0.92 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_+_173837488 | 0.90 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr8_+_85095013 | 0.89 |
ENST00000522613.1
|
RALYL
|
RALY RNA binding protein-like |
| chr8_+_38261880 | 0.88 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_1340986 | 0.87 |
ENST00000511216.1
ENST00000389851.4 |
UVSSA
|
UV-stimulated scaffold protein A |
| chr7_+_119913688 | 0.85 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr8_+_1919481 | 0.85 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr11_-_74109422 | 0.84 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr18_-_5419797 | 0.84 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_147124547 | 0.83 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr6_-_161695074 | 0.82 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_-_40657397 | 0.81 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr17_-_6735035 | 0.81 |
ENST00000338694.2
|
TEKT1
|
tektin 1 |
| chr15_+_48413211 | 0.80 |
ENST00000449382.2
|
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr1_+_230138476 | 0.78 |
ENST00000445339.1
|
RP11-552D4.1
|
RP11-552D4.1 |
| chr14_-_55658252 | 0.77 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr12_-_48119301 | 0.76 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr11_+_33061543 | 0.74 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr19_+_49622646 | 0.74 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr1_-_155939437 | 0.74 |
ENST00000609707.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_140715294 | 0.74 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
| chr8_+_30496078 | 0.73 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr1_-_85156216 | 0.73 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_-_70780770 | 0.72 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr1_+_210111534 | 0.71 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr15_+_48413169 | 0.71 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr17_-_6735012 | 0.70 |
ENST00000535086.1
|
TEKT1
|
tektin 1 |
| chr20_+_44036620 | 0.70 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_+_2014941 | 0.68 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr3_+_124303472 | 0.68 |
ENST00000291478.5
|
KALRN
|
kalirin, RhoGEF kinase |
| chr11_-_6440283 | 0.67 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr9_-_139915246 | 0.66 |
ENST00000470535.1
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr4_+_6202448 | 0.66 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr1_+_205473784 | 0.66 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr2_+_196440692 | 0.66 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_+_51863433 | 0.65 |
ENST00000444293.1
|
IQCF3
|
IQ motif containing F3 |
| chr12_+_29376592 | 0.64 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr12_+_29376673 | 0.63 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr8_-_101118439 | 0.61 |
ENST00000519408.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr4_+_62066941 | 0.61 |
ENST00000512091.2
|
LPHN3
|
latrophilin 3 |
| chrY_+_16634483 | 0.61 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr11_+_65122216 | 0.61 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr2_-_134326009 | 0.61 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr9_-_118506518 | 0.60 |
ENST00000433546.2
|
RP11-284G10.1
|
RP11-284G10.1 |
| chr12_-_48119340 | 0.60 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr9_+_6328338 | 0.60 |
ENST00000344545.5
|
TPD52L3
|
tumor protein D52-like 3 |
| chr5_-_142023766 | 0.59 |
ENST00000394496.2
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr1_+_205473720 | 0.57 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr6_-_56258892 | 0.57 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr11_-_6440624 | 0.56 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr22_+_25202232 | 0.56 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr2_-_11272234 | 0.55 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr1_+_24646263 | 0.52 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr20_+_44036900 | 0.52 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chrX_+_49593853 | 0.51 |
ENST00000376141.1
|
PAGE4
|
P antigen family, member 4 (prostate associated) |
| chr19_-_55865908 | 0.51 |
ENST00000590900.1
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr3_+_101504200 | 0.51 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr10_-_127511658 | 0.51 |
ENST00000368774.1
ENST00000368778.3 |
UROS
|
uroporphyrinogen III synthase |
| chr8_-_16859690 | 0.50 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr12_-_52911718 | 0.49 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr5_+_112074029 | 0.48 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr17_-_19281203 | 0.47 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr2_+_45878790 | 0.47 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr10_-_127511790 | 0.47 |
ENST00000368797.4
ENST00000420761.1 |
UROS
|
uroporphyrinogen III synthase |
| chrX_-_6145088 | 0.46 |
ENST00000538097.1
|
NLGN4X
|
neuroligin 4, X-linked |
| chr8_+_21914348 | 0.46 |
ENST00000520174.1
|
DMTN
|
dematin actin binding protein |
| chr2_+_96257766 | 0.46 |
ENST00000272395.2
|
TRIM43
|
tripartite motif containing 43 |
| chr17_-_8770956 | 0.45 |
ENST00000311434.9
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr7_+_37723336 | 0.45 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr8_+_28174649 | 0.44 |
ENST00000301908.3
|
PNOC
|
prepronociceptin |
| chr5_+_137225158 | 0.44 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr15_+_25200108 | 0.44 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr5_+_75379224 | 0.44 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr9_-_139094988 | 0.44 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr8_-_11324273 | 0.44 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr19_-_376011 | 0.44 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr12_+_41221975 | 0.43 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr2_-_70780572 | 0.43 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr14_+_65182395 | 0.43 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_+_24645865 | 0.42 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr2_-_133104839 | 0.42 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr17_+_66245341 | 0.42 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr12_+_123011776 | 0.42 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr1_+_24646002 | 0.41 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.40 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr2_+_181988620 | 0.39 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr3_+_33331156 | 0.39 |
ENST00000542085.1
|
FBXL2
|
F-box and leucine-rich repeat protein 2 |
| chr2_+_198669365 | 0.38 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr5_+_137225125 | 0.38 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr7_+_37723450 | 0.38 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr20_-_56286479 | 0.37 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr19_-_375970 | 0.37 |
ENST00000346878.2
|
THEG
|
theg spermatid protein |
| chr4_-_53522703 | 0.36 |
ENST00000508499.1
|
USP46
|
ubiquitin specific peptidase 46 |
| chr1_+_24645915 | 0.36 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr8_+_21914477 | 0.36 |
ENST00000517804.1
|
DMTN
|
dematin actin binding protein |
| chr14_+_29241910 | 0.36 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr5_-_137674000 | 0.36 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr12_-_54653313 | 0.35 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr10_+_92980517 | 0.34 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr15_-_31521567 | 0.34 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr14_-_25479811 | 0.34 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chrX_-_53310791 | 0.34 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chrX_-_73834449 | 0.33 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr1_-_161208013 | 0.33 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr18_+_77867177 | 0.33 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr5_+_175976324 | 0.32 |
ENST00000261944.5
|
CDHR2
|
cadherin-related family member 2 |
| chr8_-_87242589 | 0.31 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr15_+_40544749 | 0.31 |
ENST00000559617.1
ENST00000560684.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr5_+_112073544 | 0.31 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr11_-_62389577 | 0.31 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.9 | 11.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.8 | 3.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.6 | 2.4 | GO:0060168 | regulation of axon diameter(GO:0031133) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.5 | 1.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 0.9 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.4 | 1.3 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 1.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.3 | 1.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.3 | 1.3 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.3 | 1.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 1.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 2.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 1.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.3 | 1.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.3 | 3.6 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.3 | 1.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.7 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.2 | 1.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 1.4 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 2.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 4.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 3.3 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 2.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 4.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.7 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.2 | 1.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 1.2 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.2 | 1.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.9 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 7.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 2.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.3 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 2.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 2.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.7 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.8 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.4 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 1.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.8 | GO:0071205 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.9 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.6 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 2.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0033082 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 3.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 2.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 2.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 1.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.0 | 7.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.3 | 1.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 1.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 1.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 3.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 6.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.7 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 4.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 1.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.7 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 2.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 11.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.5 | 7.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 4.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 1.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.3 | 1.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.3 | 2.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 2.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 3.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 5.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 4.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 2.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 4.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.2 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 2.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 3.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 2.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 3.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 11.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 4.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 6.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |