Project
Illumina Body Map 2
Navigation
Downloads
Results for ARID5B
Z-value: 1.03
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5B | hg19_v2_chr10_+_63808970_63808990 | 0.68 | 1.7e-05 | Click! |
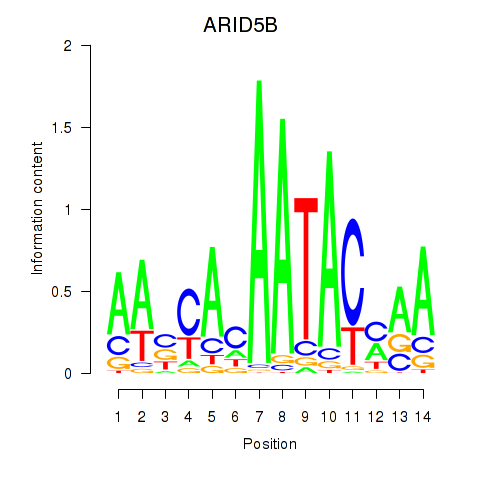
Activity profile of ARID5B motif
Sorted Z-values of ARID5B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_76995855 | 4.16 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr9_+_13446472 | 2.75 |
ENST00000428006.2
|
RP11-536O18.1
|
RP11-536O18.1 |
| chr3_+_155755482 | 2.45 |
ENST00000472028.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr8_+_19759228 | 2.44 |
ENST00000520959.1
|
LPL
|
lipoprotein lipase |
| chr7_+_36450169 | 2.14 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr14_+_32798547 | 2.03 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr14_+_32798462 | 1.87 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr17_-_3599492 | 1.86 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_3599696 | 1.84 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_10452929 | 1.80 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr11_-_111794446 | 1.66 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_63428752 | 1.62 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr18_+_32173276 | 1.56 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr12_-_8814669 | 1.55 |
ENST00000535411.1
ENST00000540087.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_8815215 | 1.54 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr11_-_102714534 | 1.49 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr12_-_8815299 | 1.46 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr1_+_86934526 | 1.44 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr10_-_98118724 | 1.43 |
ENST00000393870.2
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr10_+_53806501 | 1.40 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr12_-_122879969 | 1.39 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr22_-_22090043 | 1.38 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr15_+_51669444 | 1.37 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chrX_+_102585124 | 1.29 |
ENST00000332431.4
ENST00000372666.1 |
TCEAL7
|
transcription elongation factor A (SII)-like 7 |
| chr2_+_168675182 | 1.27 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr1_-_144995074 | 1.22 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr15_+_51669513 | 1.21 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr1_-_144994909 | 1.20 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_-_214016314 | 1.19 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr22_-_22090064 | 1.17 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chrX_+_149867681 | 1.16 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr15_+_57891609 | 1.06 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr7_+_143080063 | 1.02 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr1_-_40137710 | 1.02 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr12_-_6809958 | 1.01 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr17_-_76732928 | 1.00 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr10_-_75193308 | 0.99 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr10_-_75351088 | 0.99 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr12_-_118796971 | 0.98 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr14_+_22458631 | 0.98 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr2_-_231825668 | 0.98 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr12_-_118796910 | 0.94 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr22_+_20877924 | 0.93 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr5_-_88179017 | 0.92 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr5_+_155753745 | 0.92 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chrX_-_49089771 | 0.90 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chrX_-_11308598 | 0.89 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr11_+_1093318 | 0.89 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr3_+_68053359 | 0.82 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr4_-_44653636 | 0.82 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr12_-_10007448 | 0.82 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_48597170 | 0.81 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr10_-_29923893 | 0.81 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr5_+_161495038 | 0.79 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr14_+_23009190 | 0.79 |
ENST00000390532.1
|
TRAJ5
|
T cell receptor alpha joining 5 |
| chr4_+_95376396 | 0.78 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr10_-_128210005 | 0.78 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr6_+_155443048 | 0.77 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_+_95975672 | 0.76 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr9_+_77230499 | 0.75 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr7_+_153749732 | 0.75 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr5_+_161277603 | 0.74 |
ENST00000519621.1
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_+_44889697 | 0.73 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr1_+_78245303 | 0.72 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr1_-_144994840 | 0.72 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_-_15548591 | 0.71 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr20_+_9966728 | 0.71 |
ENST00000449270.1
|
RP5-839B4.8
|
RP5-839B4.8 |
| chr2_-_74618964 | 0.70 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr2_-_40680578 | 0.70 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_-_168698433 | 0.69 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr11_-_78052923 | 0.69 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr11_-_104769141 | 0.68 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr1_+_47264711 | 0.68 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr17_-_76713100 | 0.67 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr19_+_35849362 | 0.66 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr12_+_93096619 | 0.66 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr3_-_98241713 | 0.65 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr4_-_57524061 | 0.65 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr11_-_73882249 | 0.63 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr5_+_175288631 | 0.63 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_-_115292591 | 0.62 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr7_+_31726822 | 0.61 |
ENST00000409146.3
|
PPP1R17
|
protein phosphatase 1, regulatory subunit 17 |
| chr3_-_98241598 | 0.60 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr3_-_151102529 | 0.60 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr14_-_52436247 | 0.60 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chr1_+_104159999 | 0.60 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr4_-_159094194 | 0.59 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chrX_-_33229636 | 0.58 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr20_-_45980621 | 0.57 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_56324933 | 0.57 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr10_+_4828815 | 0.57 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr2_-_74619152 | 0.56 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr18_+_32556892 | 0.56 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_158975744 | 0.55 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr6_+_80341000 | 0.53 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_+_43855545 | 0.53 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr16_-_3422283 | 0.53 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr14_+_85994943 | 0.53 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr7_+_153749471 | 0.52 |
ENST00000406326.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr3_-_52713729 | 0.52 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr3_-_15540055 | 0.52 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr6_-_137494775 | 0.51 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chr1_+_6615241 | 0.51 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr22_+_42086547 | 0.50 |
ENST00000402966.1
|
C22orf46
|
chromosome 22 open reading frame 46 |
| chr1_+_151739131 | 0.50 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr5_+_118691706 | 0.50 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_32608566 | 0.49 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr1_-_149459549 | 0.49 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr2_+_7118755 | 0.49 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr3_-_98241760 | 0.49 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr6_+_122720681 | 0.49 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr5_+_446253 | 0.48 |
ENST00000315013.5
|
EXOC3
|
exocyst complex component 3 |
| chr12_+_56324756 | 0.48 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr19_+_38308119 | 0.48 |
ENST00000592103.1
|
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr6_+_132891461 | 0.47 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr3_+_111805182 | 0.46 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr5_-_88179302 | 0.46 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr7_-_37488777 | 0.46 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_45579762 | 0.46 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr1_+_43855560 | 0.45 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chrX_-_57164058 | 0.45 |
ENST00000374906.3
|
SPIN2A
|
spindlin family, member 2A |
| chr1_+_78245466 | 0.45 |
ENST00000477627.2
|
FAM73A
|
family with sequence similarity 73, member A |
| chr1_-_247836365 | 0.45 |
ENST00000359688.2
|
OR13G1
|
olfactory receptor, family 13, subfamily G, member 1 |
| chr17_-_36358166 | 0.44 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr1_-_224624730 | 0.44 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr1_+_248128535 | 0.44 |
ENST00000366480.3
|
OR2AK2
|
olfactory receptor, family 2, subfamily AK, member 2 |
| chr19_+_38307999 | 0.44 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr9_+_116267536 | 0.44 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr12_+_56325231 | 0.44 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr22_+_19467261 | 0.43 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr6_+_117198400 | 0.43 |
ENST00000332958.2
|
RFX6
|
regulatory factor X, 6 |
| chr12_-_11339543 | 0.42 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr1_+_87458692 | 0.42 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr3_-_98241358 | 0.42 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chrX_-_11369656 | 0.42 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr18_-_53303123 | 0.41 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr11_+_3968573 | 0.41 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr3_-_139396853 | 0.41 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr5_-_64920115 | 0.38 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr5_+_118668846 | 0.38 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr15_+_22368478 | 0.37 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr9_+_114287433 | 0.37 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr17_-_21454898 | 0.36 |
ENST00000391411.5
ENST00000412778.3 |
C17orf51
|
chromosome 17 open reading frame 51 |
| chr1_-_161337662 | 0.36 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr4_-_90229142 | 0.34 |
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3 |
| chr6_-_30080876 | 0.33 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr12_+_54366894 | 0.33 |
ENST00000546378.1
ENST00000243082.4 |
HOXC11
|
homeobox C11 |
| chr1_-_157014865 | 0.33 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr15_-_80263506 | 0.33 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr8_+_65285851 | 0.33 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr12_-_10962767 | 0.32 |
ENST00000240691.2
|
TAS2R9
|
taste receptor, type 2, member 9 |
| chr12_+_79258444 | 0.32 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr1_-_31538517 | 0.32 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr2_+_133874577 | 0.31 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr12_-_43833515 | 0.31 |
ENST00000549670.1
ENST00000395541.2 |
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr7_-_37488834 | 0.30 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_+_3379284 | 0.30 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr13_+_53216565 | 0.30 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr12_+_8309630 | 0.30 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr11_+_30253410 | 0.30 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr12_+_52281744 | 0.30 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chr4_-_25871127 | 0.28 |
ENST00000503085.1
|
RP13-494C23.1
|
RP13-494C23.1 |
| chr2_-_114300213 | 0.28 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
| chr13_-_30948036 | 0.27 |
ENST00000447147.1
ENST00000444319.1 |
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr6_+_32812568 | 0.27 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_+_126661253 | 0.27 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr20_-_9819674 | 0.26 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr8_-_116504448 | 0.26 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_+_183770835 | 0.25 |
ENST00000318351.1
|
HTR3C
|
5-hydroxytryptamine (serotonin) receptor 3C, ionotropic |
| chrX_+_106769876 | 0.25 |
ENST00000439554.1
|
FRMPD3
|
FERM and PDZ domain containing 3 |
| chr12_-_15038779 | 0.25 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr12_-_124456598 | 0.25 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chrX_+_54466829 | 0.24 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr8_-_101322132 | 0.24 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr12_+_79258547 | 0.24 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr15_+_25068773 | 0.24 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_197024394 | 0.24 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_+_43328004 | 0.24 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr5_+_154393260 | 0.24 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr3_+_161214596 | 0.23 |
ENST00000327928.4
|
OTOL1
|
otolin 1 |
| chr4_+_40201954 | 0.23 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr7_+_143792141 | 0.23 |
ENST00000408949.2
|
OR2A12
|
olfactory receptor, family 2, subfamily A, member 12 |
| chr3_+_184018352 | 0.22 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr18_+_21032781 | 0.21 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr5_+_68462837 | 0.21 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr11_-_121986923 | 0.21 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr11_-_73882057 | 0.20 |
ENST00000334126.7
ENST00000313663.7 |
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr4_-_176812842 | 0.20 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr2_+_27760247 | 0.20 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr1_-_160231451 | 0.19 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr6_-_30080863 | 0.19 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr1_+_152975488 | 0.19 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr7_-_55620433 | 0.19 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr10_-_72542272 | 0.18 |
ENST00000545575.1
|
TBATA
|
thymus, brain and testes associated |
| chr2_+_152266392 | 0.18 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr15_+_36994210 | 0.18 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr14_-_105708942 | 0.17 |
ENST00000549655.1
|
BRF1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr22_+_40573921 | 0.17 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr5_+_173763250 | 0.16 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr16_-_52061283 | 0.16 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.5 | 3.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 2.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 0.7 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.4 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 2.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 2.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 4.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 1.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.5 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 4.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 4.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.5 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.5 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 1.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 0.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 1.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.6 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 1.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.0 | GO:0046085 | purine nucleoside monophosphate catabolic process(GO:0009128) adenosine metabolic process(GO:0046085) |
| 0.1 | 1.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.7 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.0 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0009304 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 1.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 2.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 0.9 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.2 | 3.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 4.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 3.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 2.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 1.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 2.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 4.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 1.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 4.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 3.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 0.6 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.2 | 3.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 2.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 4.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 3.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |