Project
Illumina Body Map 2
Navigation
Downloads
Results for BRCA1
Z-value: 0.51
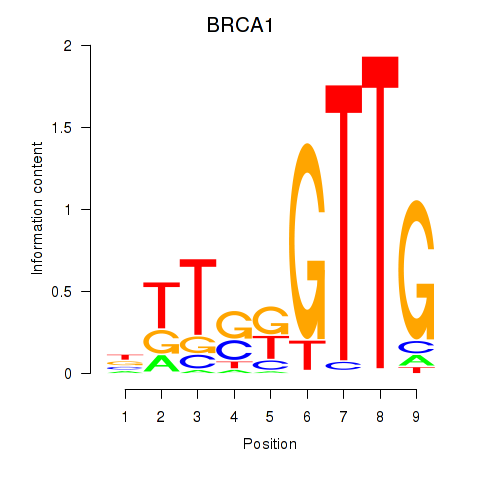
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277370_41277419 | 0.34 | 5.4e-02 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41738931 | 1.04 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr17_-_41739283 | 1.01 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr14_-_60097524 | 1.00 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr14_-_60097297 | 0.97 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr2_+_196522032 | 0.82 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_+_12717892 | 0.76 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr6_-_110797799 | 0.72 |
ENST00000456137.2
ENST00000368919.3 ENST00000439654.1 |
SLC22A16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr6_+_123100620 | 0.71 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr19_-_48059113 | 0.66 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr2_-_158182322 | 0.66 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_+_196521458 | 0.65 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr11_+_124789146 | 0.62 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr19_+_45971246 | 0.59 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr2_-_158182410 | 0.58 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr2_+_196521903 | 0.55 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr4_+_619347 | 0.54 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr17_+_74372662 | 0.51 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr4_+_619386 | 0.51 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_+_122880045 | 0.51 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr8_-_116681221 | 0.48 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_110797591 | 0.47 |
ENST00000424139.1
ENST00000437378.1 |
SLC22A16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr2_+_196521845 | 0.46 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr18_-_49557 | 0.44 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr6_-_110797642 | 0.42 |
ENST00000434949.1
|
SLC22A16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr10_-_95209 | 0.40 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr10_+_122610853 | 0.39 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr8_-_16859690 | 0.38 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr9_-_43133544 | 0.38 |
ENST00000254835.2
|
ANKRD20A3
|
ankyrin repeat domain 20 family, member A3 |
| chr10_+_104404644 | 0.35 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr2_-_158182105 | 0.35 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr14_+_22631122 | 0.34 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 (gene/pseudogene) |
| chr7_-_23510086 | 0.34 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr12_-_117319236 | 0.34 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr8_-_116681123 | 0.34 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_+_32030582 | 0.34 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr13_+_103046954 | 0.34 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr12_-_24715307 | 0.33 |
ENST00000538905.1
|
RP11-444D3.1
|
RP11-444D3.1 |
| chr10_-_95504 | 0.33 |
ENST00000447903.2
|
TUBB8
|
tubulin, beta 8 class VIII |
| chr13_-_99910673 | 0.32 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr4_-_48782259 | 0.32 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr5_-_142023766 | 0.32 |
ENST00000394496.2
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr20_+_9049682 | 0.30 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr3_+_184056614 | 0.30 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr17_-_8702667 | 0.30 |
ENST00000329805.4
|
MFSD6L
|
major facilitator superfamily domain containing 6-like |
| chrX_-_46187069 | 0.29 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_-_95510743 | 0.29 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr6_+_16129308 | 0.28 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr9_-_124976154 | 0.27 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr12_-_12491608 | 0.27 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr19_-_6057282 | 0.25 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr11_+_47291645 | 0.25 |
ENST00000395336.3
ENST00000402192.2 |
MADD
|
MAP-kinase activating death domain |
| chr11_-_71823266 | 0.24 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr19_-_40324767 | 0.24 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr12_-_67072714 | 0.24 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr3_-_176914191 | 0.23 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_+_160147176 | 0.23 |
ENST00000470705.1
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr5_+_140079919 | 0.23 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr2_-_179914760 | 0.22 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr2_-_134326009 | 0.21 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr2_-_25194476 | 0.21 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_-_78400598 | 0.21 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_+_49644470 | 0.21 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr6_+_78400375 | 0.21 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr20_-_34025999 | 0.19 |
ENST00000374369.3
|
GDF5
|
growth differentiation factor 5 |
| chr10_+_122610687 | 0.19 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr12_+_510742 | 0.19 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr5_-_68665084 | 0.19 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr1_-_173020056 | 0.18 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr13_-_99910620 | 0.18 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr12_+_510795 | 0.17 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr12_-_49449107 | 0.17 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr11_+_47291193 | 0.17 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr14_-_75179774 | 0.17 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chrX_+_10124977 | 0.17 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr7_-_78400364 | 0.17 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_140186647 | 0.17 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_-_115301235 | 0.17 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr1_-_170043709 | 0.16 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr17_-_60142609 | 0.16 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr1_+_161736072 | 0.15 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr13_-_31038370 | 0.15 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr13_-_50018241 | 0.14 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr7_+_44646177 | 0.14 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr5_+_145583107 | 0.14 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr10_-_75385711 | 0.13 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr11_-_13484713 | 0.13 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr19_+_45394477 | 0.13 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr8_+_37553261 | 0.13 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr9_-_86593238 | 0.12 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr3_+_187461442 | 0.12 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr7_+_44646162 | 0.12 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr20_-_48530230 | 0.11 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr6_+_33172407 | 0.11 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr12_-_122879969 | 0.10 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr3_+_185303962 | 0.10 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr13_-_50018140 | 0.10 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr1_+_10271674 | 0.09 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr19_-_37663332 | 0.09 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr6_-_137314371 | 0.09 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr15_+_67430339 | 0.09 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr19_-_37663572 | 0.08 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr10_+_92631709 | 0.08 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr3_-_127455200 | 0.08 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr7_+_99717230 | 0.07 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr15_-_88799948 | 0.07 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr8_+_97773457 | 0.07 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr13_+_27825706 | 0.06 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr2_+_108994466 | 0.05 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chrX_+_150565038 | 0.05 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr5_-_124082279 | 0.05 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr13_+_30002846 | 0.05 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr10_-_61900762 | 0.04 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_-_152118276 | 0.04 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr18_-_23671139 | 0.04 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr5_+_145583156 | 0.04 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr12_+_96196875 | 0.03 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr19_+_46000479 | 0.03 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr5_-_68664989 | 0.03 |
ENST00000508954.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr5_-_68665296 | 0.02 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr6_+_42981922 | 0.02 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr10_+_28822636 | 0.02 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chrX_-_70288234 | 0.02 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr13_+_30002741 | 0.02 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr5_-_68665469 | 0.02 |
ENST00000217893.5
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr11_-_110583912 | 0.01 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr7_-_99716940 | 0.01 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrX_-_77150911 | 0.01 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr12_-_49418407 | 0.01 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr8_+_97773202 | 0.00 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr7_-_6010263 | 0.00 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr1_-_217250231 | 0.00 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.3 | 2.1 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.7 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 0.5 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.4 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.6 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.3 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 1.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.3 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.7 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 1.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |