Project
Illumina Body Map 2
Navigation
Downloads
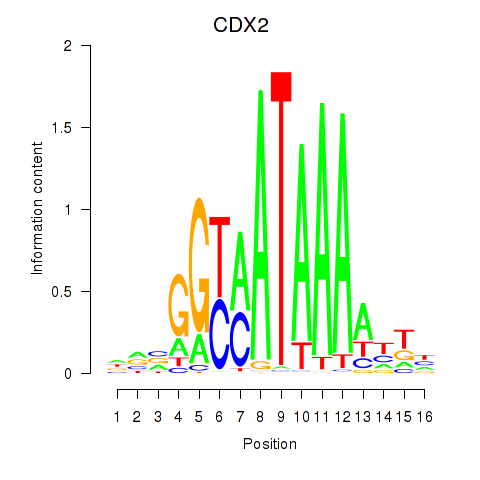
Results for CDX2
Z-value: 0.81
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX2 | hg19_v2_chr13_-_28545276_28545276 | -0.29 | 1.1e-01 | Click! |
Activity profile of CDX2 motif
Sorted Z-values of CDX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_105937068 | 1.67 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_-_237167718 | 1.66 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chrX_+_105936982 | 1.60 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr22_+_32455111 | 1.59 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr14_+_39944025 | 1.33 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_-_112443767 | 1.26 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chr10_-_94301107 | 1.26 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr12_-_112443830 | 1.26 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chrM_+_10464 | 1.17 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr10_-_103599591 | 1.14 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr17_+_58018269 | 1.11 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr6_+_111408698 | 1.07 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_+_74701062 | 0.90 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr5_+_36608280 | 0.90 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_-_72649763 | 0.89 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr10_-_24770632 | 0.88 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr22_+_17956618 | 0.88 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr7_+_138915102 | 0.88 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chrX_-_15619076 | 0.88 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr9_-_98189055 | 0.87 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr11_+_112047087 | 0.87 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr6_+_53948328 | 0.85 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr5_+_36606700 | 0.83 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_+_155484155 | 0.82 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr2_-_40657397 | 0.81 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr3_-_108672742 | 0.79 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr6_+_78400375 | 0.79 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr4_-_140222358 | 0.78 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_3663212 | 0.78 |
ENST00000397067.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr3_-_108672609 | 0.78 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr4_+_155484103 | 0.77 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr3_-_18480173 | 0.75 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr9_-_28670283 | 0.75 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr6_+_12958137 | 0.70 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr1_+_154401791 | 0.69 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr17_+_45286706 | 0.69 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr7_+_35756092 | 0.69 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr20_-_36152914 | 0.68 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr3_-_18480260 | 0.66 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chrM_+_10053 | 0.63 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_+_47666275 | 0.62 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr1_+_52682052 | 0.62 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr2_-_122262593 | 0.62 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr4_+_146539415 | 0.59 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr10_-_115614127 | 0.58 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chrX_-_30993201 | 0.58 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_+_115614370 | 0.58 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr6_+_143447322 | 0.55 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr3_+_125687987 | 0.53 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr6_+_25754927 | 0.53 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr22_+_41956767 | 0.53 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr7_+_80275663 | 0.53 |
ENST00000413265.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr20_-_7921090 | 0.51 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr20_+_42984330 | 0.50 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr7_+_35756186 | 0.50 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr10_-_129691195 | 0.50 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr16_-_66907139 | 0.50 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr2_-_51259292 | 0.50 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr4_-_169239921 | 0.50 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr18_-_52989217 | 0.50 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr13_-_50020554 | 0.49 |
ENST00000400396.1
|
AL136218.1
|
Sarcoma antigen NY-SAR-79; Uncharacterized protein |
| chr2_-_51259229 | 0.49 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr3_-_155011483 | 0.49 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr12_+_32687221 | 0.48 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_109685630 | 0.48 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr11_-_93271058 | 0.48 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr16_+_25123148 | 0.48 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr5_-_148442584 | 0.47 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr1_-_160231451 | 0.46 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr13_-_62001982 | 0.46 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr16_+_19619083 | 0.45 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chr7_+_80275621 | 0.45 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr17_-_80797886 | 0.44 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr7_-_107204918 | 0.43 |
ENST00000297135.3
|
COG5
|
component of oligomeric golgi complex 5 |
| chr7_+_80275953 | 0.43 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_+_80275752 | 0.43 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_+_60759378 | 0.41 |
ENST00000432535.1
|
LINC00844
|
long intergenic non-protein coding RNA 844 |
| chr11_+_46366918 | 0.41 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr18_+_68002675 | 0.41 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr3_-_67705006 | 0.40 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr2_+_120687335 | 0.40 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr18_+_3252206 | 0.40 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_+_12524965 | 0.40 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_-_138833630 | 0.40 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr12_+_19389814 | 0.40 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr5_+_98109322 | 0.39 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr6_+_118869452 | 0.39 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr10_+_24755416 | 0.38 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr3_+_57875711 | 0.38 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr11_+_27062860 | 0.37 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr16_+_25123041 | 0.37 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr4_-_48683188 | 0.37 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr11_+_22688150 | 0.37 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_89246647 | 0.37 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr16_+_56782118 | 0.37 |
ENST00000566678.1
|
NUP93
|
nucleoporin 93kDa |
| chr2_+_210443993 | 0.37 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_-_39646116 | 0.36 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr18_+_3252265 | 0.36 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_45686457 | 0.36 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr1_+_220267429 | 0.36 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr8_+_38662960 | 0.35 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_76871229 | 0.35 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr14_-_72458326 | 0.35 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr10_-_127505167 | 0.35 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr12_-_95774672 | 0.35 |
ENST00000549961.1
|
RP11-167N24.6
|
Uncharacterized protein |
| chr9_+_2029019 | 0.35 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_171757346 | 0.33 |
ENST00000421757.1
ENST00000415807.2 ENST00000392699.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr3_-_196911002 | 0.33 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_-_58295712 | 0.33 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr15_+_35270552 | 0.33 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr9_-_95055923 | 0.32 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr10_+_85933494 | 0.31 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr6_+_143759575 | 0.30 |
ENST00000595616.1
|
AL031320.1
|
AL031320.1 |
| chr14_+_53173910 | 0.30 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr7_+_135710494 | 0.30 |
ENST00000440744.2
|
AC024084.1
|
AC024084.1 |
| chr15_+_67835517 | 0.29 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr2_-_176046391 | 0.29 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr16_-_25122735 | 0.28 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr12_-_118796971 | 0.28 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chrX_-_135962923 | 0.27 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr10_-_5660118 | 0.27 |
ENST00000427341.1
|
RP11-336A10.4
|
RP11-336A10.4 |
| chr14_+_53173890 | 0.27 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr3_+_57875738 | 0.27 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr6_+_158733692 | 0.26 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr11_+_12302492 | 0.26 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr2_-_95787738 | 0.26 |
ENST00000272418.2
|
MRPS5
|
mitochondrial ribosomal protein S5 |
| chr9_-_130889990 | 0.26 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr9_-_73029540 | 0.25 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr11_+_22688615 | 0.25 |
ENST00000533363.1
|
GAS2
|
growth arrest-specific 2 |
| chr20_-_25320367 | 0.25 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr3_+_41241596 | 0.24 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr20_-_56647116 | 0.24 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr5_+_72143988 | 0.24 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_+_89764274 | 0.24 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr8_+_107738343 | 0.24 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr6_-_133035185 | 0.24 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr1_-_54355430 | 0.24 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr1_-_198990166 | 0.23 |
ENST00000427439.1
|
RP11-16L9.3
|
RP11-16L9.3 |
| chr15_-_41836441 | 0.23 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr2_-_8977714 | 0.23 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr13_+_24144509 | 0.23 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr9_-_37384431 | 0.23 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr9_-_95055956 | 0.23 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr12_-_10962767 | 0.23 |
ENST00000240691.2
|
TAS2R9
|
taste receptor, type 2, member 9 |
| chr4_+_95972822 | 0.23 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_+_108863651 | 0.22 |
ENST00000329106.2
ENST00000376700.1 |
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3 |
| chr20_+_2276639 | 0.22 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr19_+_21106028 | 0.21 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr12_-_76879852 | 0.21 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr6_-_26189304 | 0.21 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr8_+_85095013 | 0.21 |
ENST00000522613.1
|
RALYL
|
RALY RNA binding protein-like |
| chr3_+_15045419 | 0.21 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chrX_+_107037451 | 0.20 |
ENST00000372379.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chrX_-_135962876 | 0.20 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr15_+_86686953 | 0.20 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr5_-_148033693 | 0.20 |
ENST00000377888.3
ENST00000360693.3 |
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr2_+_210444298 | 0.19 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_+_208423840 | 0.19 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr7_+_137761167 | 0.18 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr12_+_1800179 | 0.18 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr9_-_95056010 | 0.18 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr14_+_35514323 | 0.17 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr7_+_6121296 | 0.17 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr11_+_71900703 | 0.17 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr5_-_16738451 | 0.17 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr12_+_64173583 | 0.17 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr1_+_46152886 | 0.17 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr1_+_53480598 | 0.17 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr8_+_52730143 | 0.17 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr8_-_86162077 | 0.16 |
ENST00000551479.1
|
RP11-219B4.6
|
Uncharacterized protein |
| chr12_-_72104152 | 0.16 |
ENST00000549710.1
|
RP11-498M15.1
|
RP11-498M15.1 |
| chr3_-_156534754 | 0.16 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr17_+_68071389 | 0.16 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr10_+_47657581 | 0.16 |
ENST00000441923.2
|
ANTXRL
|
anthrax toxin receptor-like |
| chr9_-_86571628 | 0.16 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr20_+_56964169 | 0.16 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_+_56546223 | 0.16 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr6_+_134274322 | 0.15 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr21_-_28338732 | 0.15 |
ENST00000284987.5
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr5_-_111093167 | 0.15 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr2_+_187371440 | 0.15 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr9_+_134065506 | 0.15 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr3_-_123710893 | 0.15 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr8_+_145215928 | 0.15 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr6_-_127840453 | 0.14 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr2_+_151485403 | 0.14 |
ENST00000413247.1
ENST00000423428.1 ENST00000427615.1 ENST00000443811.1 |
AC104777.4
|
AC104777.4 |
| chr2_+_233415363 | 0.14 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr8_-_143859197 | 0.14 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr14_+_52456327 | 0.13 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr7_-_111032971 | 0.13 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_65996599 | 0.13 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr2_+_208423891 | 0.13 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr4_+_26165074 | 0.13 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_56546363 | 0.13 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr4_+_70894130 | 0.13 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr12_-_118796910 | 0.13 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr16_+_66613351 | 0.13 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr19_-_53636125 | 0.12 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr2_+_109204743 | 0.12 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_13885252 | 0.12 |
ENST00000221576.4
|
C19orf53
|
chromosome 19 open reading frame 53 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.3 | 0.8 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.3 | 1.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 3.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 0.8 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 1.6 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.2 | 0.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 1.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.7 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.8 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.6 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 1.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.1 | 1.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.3 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.4 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.2 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.2 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.6 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) vascular wound healing(GO:0061042) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 2.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.3 | 0.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 0.8 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.3 | 1.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 1.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.7 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.5 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.2 | 0.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 1.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.8 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 1.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |