Project
Illumina Body Map 2
Navigation
Downloads
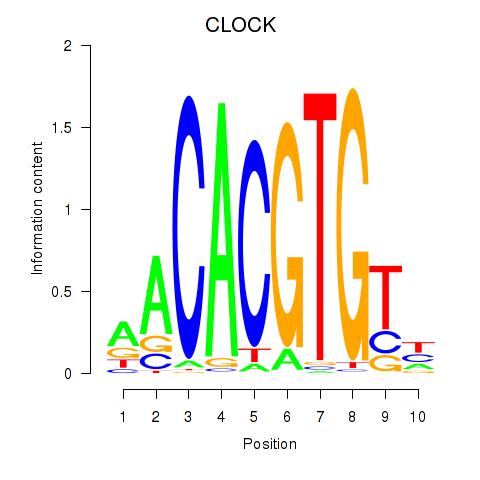
Results for CLOCK
Z-value: 1.57
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | clock circadian regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CLOCK | hg19_v2_chr4_-_56412713_56412799 | -0.30 | 9.0e-02 | Click! |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_23877474 | 5.07 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr14_-_23876801 | 4.90 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr16_+_56703703 | 4.26 |
ENST00000332374.4
|
MT1H
|
metallothionein 1H |
| chr18_-_3220106 | 3.79 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr16_+_56703737 | 3.58 |
ENST00000569155.1
|
MT1H
|
metallothionein 1H |
| chr12_-_103310987 | 3.33 |
ENST00000307000.2
|
PAH
|
phenylalanine hydroxylase |
| chr1_-_40137710 | 3.21 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr16_-_81129845 | 3.08 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr22_+_18593097 | 2.85 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr4_+_74275057 | 2.57 |
ENST00000511370.1
|
ALB
|
albumin |
| chr6_+_73331918 | 2.44 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr1_+_100315613 | 2.16 |
ENST00000361915.3
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr18_-_3219847 | 2.13 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr5_+_70883154 | 2.13 |
ENST00000509358.2
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr17_+_72428266 | 2.11 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_72929319 | 2.09 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr17_-_9694614 | 2.08 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr16_+_14980632 | 2.05 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr22_+_18593446 | 2.03 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr2_-_220118631 | 2.02 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr16_+_6533380 | 2.01 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr10_-_75415825 | 2.00 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr11_-_119252359 | 1.99 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr17_+_72428218 | 1.97 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_+_8814563 | 1.96 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr22_+_18593507 | 1.95 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr7_+_116312411 | 1.95 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr11_-_59951955 | 1.92 |
ENST00000531531.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr11_+_72929402 | 1.90 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr1_-_15911510 | 1.89 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr5_+_70883117 | 1.87 |
ENST00000340941.6
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chrX_+_10124977 | 1.86 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr1_+_150254936 | 1.85 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr5_+_70883178 | 1.84 |
ENST00000323375.8
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr1_-_19229218 | 1.81 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_19229248 | 1.81 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_+_64854958 | 1.81 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr16_-_46782221 | 1.79 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr1_-_19229014 | 1.77 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr3_+_35682913 | 1.77 |
ENST00000449196.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_119252425 | 1.76 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr4_-_186661365 | 1.75 |
ENST00000452351.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_71833756 | 1.73 |
ENST00000536515.1
ENST00000540815.2 |
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr14_-_73360796 | 1.70 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr5_+_43602750 | 1.70 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr4_-_46996424 | 1.68 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr12_-_49393092 | 1.67 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr9_-_135545380 | 1.66 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr2_+_17721937 | 1.65 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr2_+_17721230 | 1.65 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr8_+_95908041 | 1.65 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr17_+_48172639 | 1.64 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr9_+_135545409 | 1.64 |
ENST00000483873.2
ENST00000372146.4 |
GTF3C4
|
general transcription factor IIIC, polypeptide 4, 90kDa |
| chr17_-_7017559 | 1.63 |
ENST00000446679.2
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr4_-_185729602 | 1.63 |
ENST00000437665.3
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_-_146057144 | 1.61 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr10_+_104486253 | 1.59 |
ENST00000602868.1
|
SFXN2
|
sideroflexin 2 |
| chr10_-_5708515 | 1.57 |
ENST00000357700.6
|
ASB13
|
ankyrin repeat and SOCS box containing 13 |
| chr11_+_45792967 | 1.56 |
ENST00000378779.2
|
CTD-2210P24.4
|
Uncharacterized protein |
| chr15_+_36871806 | 1.56 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr12_+_57624119 | 1.56 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_+_43603229 | 1.55 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr7_+_75677354 | 1.54 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr12_+_56661461 | 1.53 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr5_+_96077888 | 1.52 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr2_-_71454185 | 1.52 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr12_+_57624085 | 1.52 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_-_59951889 | 1.51 |
ENST00000532169.1
ENST00000534596.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr1_+_62439037 | 1.51 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr10_-_69597810 | 1.48 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr15_+_91446961 | 1.44 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr3_-_125899721 | 1.42 |
ENST00000488356.1
|
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr12_+_104235229 | 1.41 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr17_+_72427477 | 1.41 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_17721920 | 1.39 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr1_+_111991474 | 1.39 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr3_+_186358200 | 1.38 |
ENST00000382136.3
|
FETUB
|
fetuin B |
| chr16_+_89238149 | 1.38 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr7_+_75677465 | 1.38 |
ENST00000432020.2
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr9_-_130890662 | 1.38 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chr17_-_2615031 | 1.37 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr8_+_95907993 | 1.37 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr3_+_72201910 | 1.36 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr4_+_75023816 | 1.35 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr21_+_38071430 | 1.34 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr21_-_16374688 | 1.33 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr3_-_71777824 | 1.31 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr1_-_231376836 | 1.30 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr1_-_216978709 | 1.28 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr12_+_57624059 | 1.28 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_-_220117867 | 1.27 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr15_-_52944231 | 1.26 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr15_+_36871983 | 1.26 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr6_+_123110465 | 1.25 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr8_+_21915368 | 1.24 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr10_-_120925054 | 1.23 |
ENST00000419372.1
ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4
|
sideroflexin 4 |
| chr10_-_69597915 | 1.22 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_+_154955797 | 1.22 |
ENST00000368433.1
ENST00000315144.10 ENST00000368432.1 ENST00000368431.3 ENST00000292180.3 |
FLAD1
|
flavin adenine dinucleotide synthetase 1 |
| chr17_-_17942473 | 1.21 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr18_+_72163536 | 1.20 |
ENST00000579847.1
ENST00000583203.1 ENST00000581513.1 ENST00000577600.1 ENST00000579583.1 ENST00000584613.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_+_72052964 | 1.20 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr3_-_50329835 | 1.19 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr12_+_81110684 | 1.18 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr17_-_2614927 | 1.17 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr18_+_72163443 | 1.17 |
ENST00000324262.4
ENST00000580672.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr12_+_113623325 | 1.17 |
ENST00000549621.1
ENST00000548278.1 ENST00000552495.1 |
C12orf52
|
RBPJ interacting and tubulin associated 1 |
| chr11_-_59952106 | 1.16 |
ENST00000529054.1
ENST00000530839.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr10_+_123970670 | 1.15 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_+_57623869 | 1.15 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_-_62332357 | 1.15 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_-_124553437 | 1.15 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr3_-_188665428 | 1.13 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr1_-_11986442 | 1.10 |
ENST00000376572.3
ENST00000376576.3 |
KIAA2013
|
KIAA2013 |
| chr17_-_71088797 | 1.10 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr3_+_183967409 | 1.10 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr12_+_57623477 | 1.10 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr16_-_4588822 | 1.10 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr10_+_60028818 | 1.10 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr3_-_125899645 | 1.09 |
ENST00000393434.2
ENST00000460368.1 |
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr18_+_43913919 | 1.09 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr19_+_13001840 | 1.08 |
ENST00000222214.5
ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH
|
glutaryl-CoA dehydrogenase |
| chr3_-_50329990 | 1.08 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr1_-_113498616 | 1.08 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_-_45307817 | 1.06 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr17_+_55163075 | 1.06 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr9_+_706842 | 1.05 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr17_+_46970178 | 1.05 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_+_57623907 | 1.05 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr3_+_184032919 | 1.04 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_+_149535022 | 1.04 |
ENST00000466795.1
|
RNF13
|
ring finger protein 13 |
| chr5_+_110074685 | 1.04 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr1_-_231376867 | 1.03 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr20_+_57430162 | 1.03 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr9_+_34646651 | 1.02 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr3_-_125899414 | 1.02 |
ENST00000393431.2
ENST00000452905.2 ENST00000455064.2 |
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr15_+_49447947 | 1.02 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr11_+_22688615 | 1.01 |
ENST00000533363.1
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_231376941 | 1.01 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr16_-_4588762 | 1.00 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr14_-_68066849 | 1.00 |
ENST00000558493.1
ENST00000561272.1 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr5_+_176784837 | 1.00 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr3_+_52720187 | 0.99 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr17_+_21279509 | 0.99 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr1_+_11333245 | 0.99 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr11_-_61101247 | 0.98 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chrX_-_63425561 | 0.98 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr2_-_47143160 | 0.98 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr6_+_123110302 | 0.98 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr17_+_46970134 | 0.97 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr11_+_18416133 | 0.97 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr7_+_95115210 | 0.97 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr10_+_180405 | 0.96 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr7_-_47621229 | 0.96 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr9_-_6015607 | 0.96 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr16_+_771663 | 0.96 |
ENST00000568916.1
|
FAM173A
|
family with sequence similarity 173, member A |
| chr19_+_41903709 | 0.95 |
ENST00000542943.1
ENST00000457836.2 |
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr6_-_146056341 | 0.95 |
ENST00000435470.1
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr14_-_80677815 | 0.94 |
ENST00000557125.1
ENST00000555750.1 |
DIO2
|
deiodinase, iodothyronine, type II |
| chr7_-_148725544 | 0.94 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr7_+_56019486 | 0.94 |
ENST00000446692.1
ENST00000285298.4 ENST00000443449.1 |
GBAS
MRPS17
|
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr17_+_46970127 | 0.94 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr14_+_32030582 | 0.94 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr3_-_183543301 | 0.94 |
ENST00000318631.3
ENST00000431348.1 |
MAP6D1
|
MAP6 domain containing 1 |
| chr9_-_136203235 | 0.94 |
ENST00000372022.4
|
SURF6
|
surfeit 6 |
| chr1_+_67773527 | 0.93 |
ENST00000541374.1
ENST00000544434.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr14_+_39736582 | 0.93 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr2_+_216176761 | 0.91 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr8_-_110656995 | 0.90 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr2_+_68384976 | 0.90 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr8_+_67104940 | 0.90 |
ENST00000517689.1
|
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr1_+_26496362 | 0.90 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr22_+_24990746 | 0.89 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr11_+_18416103 | 0.89 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr9_+_34646624 | 0.89 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr6_-_31974881 | 0.88 |
ENST00000594256.1
|
AL645922.1
|
Uncharacterized protein |
| chr22_-_30987837 | 0.88 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr7_-_103629963 | 0.88 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr4_-_54457783 | 0.88 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr2_-_74618907 | 0.87 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr12_+_6644443 | 0.87 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr14_-_80677613 | 0.86 |
ENST00000556811.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr18_+_33877654 | 0.86 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr11_-_47470703 | 0.85 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr1_-_111991850 | 0.85 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr9_+_96051469 | 0.85 |
ENST00000453718.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr3_-_167813672 | 0.85 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr3_-_53878644 | 0.84 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr11_-_47664072 | 0.84 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr16_-_18937072 | 0.84 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr10_-_69597828 | 0.84 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_205744205 | 0.83 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr7_-_44163107 | 0.82 |
ENST00000406581.2
ENST00000452185.1 ENST00000436844.1 ENST00000418438.1 |
POLD2
|
polymerase (DNA directed), delta 2, accessory subunit |
| chr3_-_183967296 | 0.82 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr2_-_47142884 | 0.81 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_-_5342380 | 0.81 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr1_-_197744763 | 0.80 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr6_-_97345689 | 0.80 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr12_+_6833323 | 0.80 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr19_+_34895289 | 0.80 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr1_-_35325318 | 0.79 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.9 | 5.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.9 | 2.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 7.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.7 | 3.5 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.7 | 10.5 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.6 | 1.9 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.6 | 1.8 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.6 | 3.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 1.8 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.6 | 5.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.5 | 3.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.5 | 3.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.4 | 1.3 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.4 | 1.7 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.4 | 1.2 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.4 | 2.0 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.4 | 1.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.4 | 1.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 2.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 1.6 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 0.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 4.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.3 | 1.6 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 1.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.3 | 1.5 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.3 | 1.3 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.3 | 1.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.2 | 7.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 0.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 1.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.2 | 0.7 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.6 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.2 | 0.8 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 1.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 0.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.6 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.2 | 1.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 1.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 2.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 2.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 1.3 | GO:0097319 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 0.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.2 | 1.7 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 0.5 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 1.6 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 2.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.7 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 3.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.5 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.2 | 3.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 2.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 3.3 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 3.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 1.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 2.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 1.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 1.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 0.6 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.2 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 2.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 1.6 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 5.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 1.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.5 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 2.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 1.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.7 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.6 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.6 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 2.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.6 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.6 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 1.0 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.6 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 3.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.3 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.7 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.3 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 1.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 1.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.2 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 0.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 0.2 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.2 | GO:0052501 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 4.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.6 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.5 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 1.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.5 | GO:0014870 | response to muscle inactivity(GO:0014870) |
| 0.1 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.3 | GO:0090526 | positive regulation of glucokinase activity(GO:0033133) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.5 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.1 | 0.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.6 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 1.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.7 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.0 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 5.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.3 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.6 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 2.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 2.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.6 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.6 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.3 | GO:1902953 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.5 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 1.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 2.1 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 1.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 1.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.6 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.8 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 1.8 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 2.4 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 3.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.6 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0071874 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.8 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 2.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.7 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 1.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0006721 | retinoid metabolic process(GO:0001523) isoprenoid metabolic process(GO:0006720) terpenoid metabolic process(GO:0006721) diterpenoid metabolic process(GO:0016101) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:2000672 | regulation of motor neuron apoptotic process(GO:2000671) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 1.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.9 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.1 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.6 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 1.0 | 3.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 1.0 | 5.8 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.8 | 7.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.6 | 2.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 2.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 9.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 1.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 1.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.3 | 5.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 2.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 1.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 1.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.3 | GO:0030684 | preribosome(GO:0030684) |
| 0.1 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 2.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.5 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 2.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 1.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 4.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 3.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 15.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 3.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 5.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 10.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 1.1 | 10.0 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.1 | 3.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 1.1 | 3.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 5.8 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.8 | 3.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.8 | 7.7 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.7 | 3.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.6 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 3.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 2.4 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.5 | 1.4 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.4 | 2.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.4 | 2.0 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 1.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.3 | 1.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.3 | 1.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 1.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.3 | 2.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 1.0 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.3 | 0.9 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.3 | 2.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 1.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 0.8 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 1.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.2 | 1.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 0.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.6 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.2 | 0.6 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.2 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 6.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.8 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 1.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 0.5 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.2 | 5.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 2.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 0.5 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.2 | 0.6 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 1.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 2.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 2.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 4.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.9 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 1.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 2.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 2.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.6 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.3 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 3.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.5 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 1.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 12.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.6 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 2.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 1.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.9 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.7 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 1.6 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 4.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 6.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 5.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 5.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 4.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 9.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 10.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |