Project
Illumina Body Map 2
Navigation
Downloads
Results for CREB3L1_CREB3
Z-value: 0.87
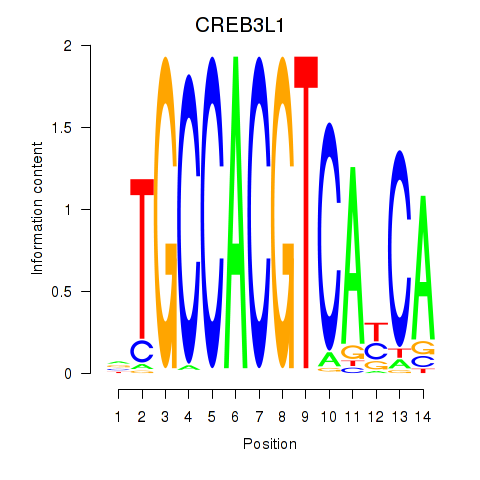
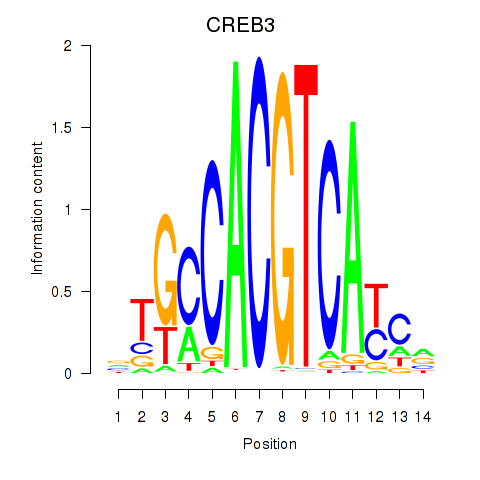
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L1 | hg19_v2_chr11_+_46332679_46332708 | -0.19 | 3.0e-01 | Click! |
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | -0.15 | 4.2e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_38864041 | 4.40 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr6_+_31916733 | 3.98 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr3_+_100211412 | 3.35 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr14_+_24600484 | 2.40 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr16_+_2083265 | 1.75 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr12_-_105352047 | 1.63 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr16_-_68269971 | 1.59 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr7_-_73184588 | 1.59 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr9_+_124461603 | 1.53 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein |
| chr19_+_50031547 | 1.45 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr1_+_119911425 | 1.42 |
ENST00000361035.4
ENST00000325945.3 |
HAO2
|
hydroxyacid oxidase 2 (long chain) |
| chr7_-_100493744 | 1.29 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr7_+_94023873 | 1.21 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr9_+_101984577 | 1.20 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr1_+_119911396 | 1.20 |
ENST00000457318.1
|
HAO2
|
hydroxyacid oxidase 2 (long chain) |
| chr12_+_56624436 | 1.18 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr16_-_11836595 | 1.16 |
ENST00000356957.3
ENST00000283033.5 |
TXNDC11
|
thioredoxin domain containing 11 |
| chrX_+_153060090 | 1.13 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr13_-_45915221 | 1.12 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr7_+_16793160 | 1.05 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chrX_+_153059608 | 1.05 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr14_-_35344093 | 1.02 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr19_-_48894104 | 1.01 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr4_+_75311019 | 1.01 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr1_-_155145721 | 1.00 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr17_-_191188 | 0.99 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr12_-_105352080 | 0.96 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr2_-_69614373 | 0.91 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr2_+_27255806 | 0.90 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr8_-_38126635 | 0.90 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr22_-_28392227 | 0.88 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr9_+_100818976 | 0.85 |
ENST00000210444.5
|
NANS
|
N-acetylneuraminic acid synthase |
| chr2_-_27341966 | 0.84 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr19_-_4670345 | 0.84 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr8_-_38126675 | 0.84 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr12_-_120638902 | 0.83 |
ENST00000551150.1
ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0
|
ribosomal protein, large, P0 |
| chr19_-_3985455 | 0.83 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr4_+_75310851 | 0.80 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr9_-_127703333 | 0.79 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr11_-_67407031 | 0.78 |
ENST00000335385.3
|
TBX10
|
T-box 10 |
| chr1_-_119683251 | 0.77 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr3_-_132378919 | 0.77 |
ENST00000355458.3
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr11_+_65292538 | 0.76 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr12_-_106641728 | 0.76 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr3_-_132378897 | 0.75 |
ENST00000545291.1
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr3_-_57583052 | 0.74 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr15_+_73976715 | 0.72 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr9_-_136242956 | 0.71 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr3_-_57583130 | 0.71 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr1_+_222913009 | 0.71 |
ENST00000456298.1
|
FAM177B
|
family with sequence similarity 177, member B |
| chr16_+_2059872 | 0.70 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr3_+_133292574 | 0.70 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr5_+_133984462 | 0.70 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr19_+_50354462 | 0.70 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr6_+_30908747 | 0.69 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr1_-_119682812 | 0.68 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr9_+_114393581 | 0.68 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr11_-_62341445 | 0.68 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr4_+_25235597 | 0.68 |
ENST00000264864.6
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr19_+_5681153 | 0.67 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr4_+_128982416 | 0.67 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr1_+_162531294 | 0.67 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr11_+_69455855 | 0.67 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr8_+_22423168 | 0.65 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_+_222791417 | 0.62 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr9_-_101984184 | 0.62 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr4_+_128982490 | 0.62 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chrX_+_47441712 | 0.61 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr9_-_98079965 | 0.61 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr4_-_111544254 | 0.61 |
ENST00000306732.3
|
PITX2
|
paired-like homeodomain 2 |
| chr16_+_56642489 | 0.61 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr19_+_50354430 | 0.61 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr11_-_59383617 | 0.61 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr1_+_170632250 | 0.61 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr16_+_19125252 | 0.60 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr8_+_140943416 | 0.60 |
ENST00000507535.3
|
C8orf17
|
chromosome 8 open reading frame 17 |
| chr11_+_65292884 | 0.60 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr17_+_40688190 | 0.60 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr11_-_118927561 | 0.59 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr11_-_118927816 | 0.59 |
ENST00000534233.1
ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1
|
hypoxia up-regulated 1 |
| chr8_+_22423219 | 0.58 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_-_113909877 | 0.57 |
ENST00000261731.3
|
LHX5
|
LIM homeobox 5 |
| chr10_+_64893039 | 0.57 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr17_-_66951474 | 0.57 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr12_-_49318715 | 0.56 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr16_+_56642041 | 0.56 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr2_-_27341765 | 0.55 |
ENST00000405600.1
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr2_-_242211359 | 0.54 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr17_+_7211656 | 0.53 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr4_+_128982430 | 0.53 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr20_+_31595406 | 0.53 |
ENST00000170150.3
|
BPIFB2
|
BPI fold containing family B, member 2 |
| chr4_-_119757239 | 0.52 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr8_+_22423479 | 0.52 |
ENST00000522721.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr8_-_64080945 | 0.52 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr3_+_122785895 | 0.52 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr11_-_118927735 | 0.51 |
ENST00000526656.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr8_+_22422749 | 0.50 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr11_-_62599505 | 0.50 |
ENST00000377897.4
ENST00000394690.1 ENST00000541317.1 ENST00000294179.3 |
STX5
|
syntaxin 5 |
| chr2_+_183580954 | 0.49 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr4_-_111544207 | 0.48 |
ENST00000557119.2
|
PITX2
|
paired-like homeodomain 2 |
| chr9_+_139377947 | 0.48 |
ENST00000354376.1
|
C9orf163
|
chromosome 9 open reading frame 163 |
| chr11_+_32112431 | 0.47 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr1_+_44445549 | 0.47 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr10_-_22292675 | 0.47 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr7_+_72848092 | 0.46 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr20_-_17662705 | 0.46 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr12_+_107349497 | 0.46 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr11_-_118927880 | 0.45 |
ENST00000527038.2
|
HYOU1
|
hypoxia up-regulated 1 |
| chr6_+_36164487 | 0.44 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr12_-_56123444 | 0.44 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr17_-_66951382 | 0.43 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr10_-_105437909 | 0.43 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr3_-_49395705 | 0.43 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr6_-_31938700 | 0.43 |
ENST00000495340.1
|
DXO
|
decapping exoribonuclease |
| chr11_-_68039364 | 0.43 |
ENST00000533310.1
ENST00000304271.6 ENST00000527280.1 |
C11orf24
|
chromosome 11 open reading frame 24 |
| chr2_-_43823093 | 0.42 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr9_-_130477912 | 0.42 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr11_-_67276100 | 0.42 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr3_+_128598433 | 0.42 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr11_-_118972575 | 0.42 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr1_+_44445643 | 0.41 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_-_2059748 | 0.41 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr19_+_50354393 | 0.41 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr20_-_17662878 | 0.41 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr2_-_114514181 | 0.40 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr16_-_66959429 | 0.40 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr12_+_107349606 | 0.40 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chrX_+_48660287 | 0.39 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr6_+_151561085 | 0.38 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_-_56321649 | 0.38 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr5_+_94890778 | 0.38 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr19_-_46088068 | 0.38 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr5_+_109025067 | 0.37 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr10_-_30348439 | 0.37 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr12_-_96336369 | 0.36 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr5_-_39424961 | 0.35 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_100860949 | 0.35 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr7_+_6522922 | 0.35 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr5_+_138210919 | 0.35 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr19_+_50979753 | 0.34 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr13_-_52027134 | 0.34 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr5_+_65440032 | 0.34 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr3_-_156272872 | 0.34 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr16_-_73082274 | 0.34 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr3_+_47422485 | 0.34 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr9_-_74980113 | 0.33 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr14_-_50053081 | 0.33 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr4_+_141445333 | 0.33 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr1_-_206945830 | 0.32 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr11_+_809961 | 0.32 |
ENST00000530797.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr5_-_131562935 | 0.32 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr8_-_71519889 | 0.32 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr5_-_32174369 | 0.32 |
ENST00000265070.6
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein) |
| chr12_-_32040123 | 0.32 |
ENST00000535163.1
|
RP11-428G5.5
|
RP11-428G5.5 |
| chr8_-_57906362 | 0.32 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr2_-_43823119 | 0.32 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr5_-_39425290 | 0.31 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_+_6554971 | 0.31 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr17_-_46716647 | 0.31 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr16_-_10868853 | 0.30 |
ENST00000572428.1
|
TVP23A
|
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chr14_-_50087312 | 0.30 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr12_-_56320887 | 0.30 |
ENST00000398213.4
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr19_-_10341948 | 0.30 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr21_-_38445297 | 0.30 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr5_-_39425222 | 0.30 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_100271355 | 0.30 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr3_-_49395892 | 0.30 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr12_+_56110247 | 0.30 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr2_+_192542879 | 0.29 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_48450575 | 0.29 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr7_+_150690857 | 0.29 |
ENST00000484524.1
ENST00000467517.1 |
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr11_-_3818932 | 0.29 |
ENST00000324932.7
ENST00000359171.4 |
NUP98
|
nucleoporin 98kDa |
| chr3_-_113465065 | 0.29 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr17_-_77967433 | 0.29 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_153940097 | 0.29 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr5_-_1799932 | 0.28 |
ENST00000382647.7
ENST00000505059.2 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr13_+_27998681 | 0.28 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr16_-_69390209 | 0.28 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr5_-_1799965 | 0.27 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr17_-_7155274 | 0.27 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chrX_-_10544942 | 0.27 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr19_+_50353944 | 0.27 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr1_+_228270361 | 0.26 |
ENST00000272102.5
ENST00000540651.1 |
ARF1
|
ADP-ribosylation factor 1 |
| chr4_-_83350580 | 0.26 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr17_+_7155343 | 0.26 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr5_+_149340339 | 0.25 |
ENST00000433184.1
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr1_+_228270784 | 0.25 |
ENST00000541182.1
|
ARF1
|
ADP-ribosylation factor 1 |
| chr19_+_531713 | 0.25 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr12_-_10766184 | 0.25 |
ENST00000539554.1
ENST00000381881.2 ENST00000320756.2 |
MAGOHB
|
mago-nashi homolog B (Drosophila) |
| chr14_+_50087468 | 0.25 |
ENST00000305386.2
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr8_+_64081118 | 0.25 |
ENST00000539294.1
|
YTHDF3
|
YTH domain family, member 3 |
| chr19_-_5680891 | 0.25 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr2_+_96068436 | 0.24 |
ENST00000445649.1
ENST00000447036.1 ENST00000233379.4 ENST00000418606.1 |
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr11_-_64889649 | 0.24 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr1_+_32645269 | 0.24 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr12_-_50616382 | 0.24 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr5_+_94890840 | 0.24 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chrX_-_153237258 | 0.24 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr8_-_29208183 | 0.23 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr17_+_33307503 | 0.23 |
ENST00000378526.4
ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3
|
ligase III, DNA, ATP-dependent |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.4 | 5.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.5 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.3 | 2.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 1.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 1.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 1.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.7 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.2 | 1.8 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 0.5 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.2 | 4.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.5 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.7 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 2.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 1.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.9 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.8 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 1.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.2 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.2 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.8 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.4 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 1.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.8 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 1.0 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 1.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 2.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 3.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 2.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 2.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 1.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.6 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.2 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 3.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 1.2 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 0.2 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 2.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 2.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 4.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.6 | 5.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 1.5 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 1.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 1.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 2.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 4.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 1.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.6 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 3.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.0 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 4.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 4.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 7.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 7.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |