Project
Illumina Body Map 2
Navigation
Downloads
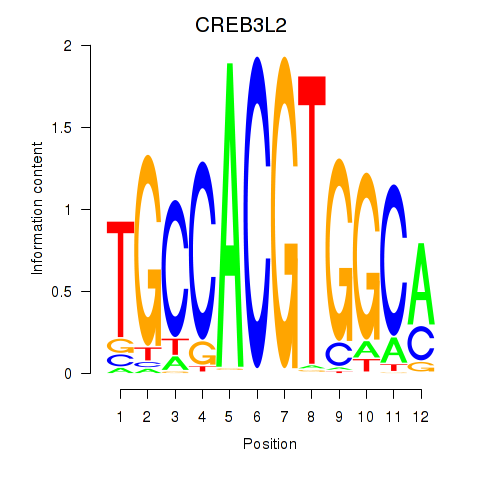
Results for CREB3L2
Z-value: 1.17
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | cAMP responsive element binding protein 3 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg19_v2_chr7_-_137686791_137686821 | -0.02 | 8.9e-01 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_61584026 | 4.06 |
ENST00000370351.4
ENST00000370349.3 |
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr20_+_61584398 | 3.39 |
ENST00000411611.1
|
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr19_-_4670345 | 2.49 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr20_-_22559211 | 2.39 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr17_-_26695013 | 2.27 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr17_-_26694979 | 2.06 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr9_+_96928516 | 1.83 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr12_-_49318715 | 1.83 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr14_+_100789669 | 1.81 |
ENST00000361529.3
ENST00000557052.1 |
SLC25A47
|
solute carrier family 25, member 47 |
| chr5_+_34656569 | 1.76 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr5_+_34656529 | 1.66 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr1_-_32801825 | 1.64 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr5_+_34656331 | 1.60 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr20_-_61051026 | 1.59 |
ENST00000252997.2
|
GATA5
|
GATA binding protein 5 |
| chr10_+_102106829 | 1.54 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr7_-_100171270 | 1.54 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr14_-_106174960 | 1.52 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr14_-_106054659 | 1.44 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr20_+_31595406 | 1.41 |
ENST00000170150.3
|
BPIFB2
|
BPI fold containing family B, member 2 |
| chr10_+_23216944 | 1.41 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chr8_+_22225041 | 1.41 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr18_+_56806701 | 1.40 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr2_+_217498105 | 1.39 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr22_+_22681656 | 1.39 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr9_+_130478345 | 1.38 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr3_+_122785895 | 1.36 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr8_+_22224760 | 1.32 |
ENST00000359741.5
ENST00000520644.1 ENST00000240095.6 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr19_-_11688500 | 1.32 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr20_-_2821271 | 1.31 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr5_-_138725594 | 1.30 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr18_+_56807096 | 1.30 |
ENST00000588875.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr18_+_9708162 | 1.29 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr19_-_42759300 | 1.26 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr15_+_89181974 | 1.25 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_+_151561506 | 1.25 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr10_+_23217006 | 1.24 |
ENST00000376528.4
ENST00000447081.1 |
ARMC3
|
armadillo repeat containing 3 |
| chr17_+_72270429 | 1.24 |
ENST00000311014.6
|
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr2_-_85108164 | 1.20 |
ENST00000409520.2
|
TRABD2A
|
TraB domain containing 2A |
| chr22_+_44568825 | 1.20 |
ENST00000422871.1
|
PARVG
|
parvin, gamma |
| chr5_-_138725560 | 1.20 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr18_-_3013307 | 1.20 |
ENST00000584294.1
|
LPIN2
|
lipin 2 |
| chr22_+_38864041 | 1.16 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr14_+_101293687 | 1.16 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr15_+_58724184 | 1.15 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr10_+_112257596 | 1.14 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr2_+_198365095 | 1.12 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr15_+_89182156 | 1.12 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_+_89419370 | 1.11 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr21_+_45875354 | 1.06 |
ENST00000291592.4
|
LRRC3
|
leucine rich repeat containing 3 |
| chr3_+_184098065 | 1.06 |
ENST00000348986.3
|
CHRD
|
chordin |
| chr5_+_34656450 | 1.06 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr2_-_85108363 | 1.05 |
ENST00000335459.5
|
TRABD2A
|
TraB domain containing 2A |
| chr6_+_151561085 | 1.05 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_-_85108240 | 1.05 |
ENST00000409133.1
|
TRABD2A
|
TraB domain containing 2A |
| chr19_-_47164386 | 1.02 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr5_+_176730769 | 1.01 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr1_-_27240455 | 0.99 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr6_-_7911042 | 0.98 |
ENST00000379757.4
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr15_+_89182178 | 0.96 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr4_+_37455536 | 0.96 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr6_-_136871957 | 0.96 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr17_+_48450575 | 0.95 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr7_+_29603394 | 0.95 |
ENST00000319694.2
|
PRR15
|
proline rich 15 |
| chr11_+_120081475 | 0.94 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr1_+_1567474 | 0.93 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B |
| chr3_-_123603137 | 0.91 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr7_-_6523755 | 0.89 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr3_-_178789220 | 0.89 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr21_-_18985158 | 0.85 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr1_-_42921915 | 0.83 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr15_-_52404921 | 0.82 |
ENST00000561198.1
ENST00000260442.3 |
BCL2L10
|
BCL2-like 10 (apoptosis facilitator) |
| chr15_+_62359175 | 0.82 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium-dependent domain containing 4A |
| chr21_-_40720995 | 0.81 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr1_-_241520525 | 0.80 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr8_+_56014949 | 0.80 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr19_+_49458107 | 0.78 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr12_+_121416489 | 0.78 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr21_-_18985230 | 0.78 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr6_-_32811771 | 0.76 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr20_-_4795747 | 0.76 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr7_+_2671568 | 0.72 |
ENST00000258796.7
|
TTYH3
|
tweety family member 3 |
| chr19_+_12305895 | 0.72 |
ENST00000451691.2
|
CTD-2666L21.1
|
CTD-2666L21.1 |
| chr8_-_99129338 | 0.72 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr16_+_215965 | 0.72 |
ENST00000356815.3
|
HBM
|
hemoglobin, mu |
| chrX_+_128913906 | 0.72 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr19_-_11688951 | 0.71 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr12_+_121416340 | 0.70 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr21_-_40720974 | 0.70 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr16_-_67185117 | 0.69 |
ENST00000449549.3
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr19_+_51774540 | 0.69 |
ENST00000600813.1
|
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr1_+_113392455 | 0.69 |
ENST00000456651.1
ENST00000422022.1 |
RP3-522D1.1
|
RP3-522D1.1 |
| chr2_-_198364581 | 0.68 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr13_+_98794810 | 0.68 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr8_-_99129384 | 0.68 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr1_-_11865982 | 0.67 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_+_11866207 | 0.66 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr3_-_57583185 | 0.66 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr7_+_100464760 | 0.66 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr1_-_91317072 | 0.65 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr1_+_28918712 | 0.65 |
ENST00000373826.3
|
RAB42
|
RAB42, member RAS oncogene family |
| chr17_+_74723031 | 0.64 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr7_-_121036337 | 0.64 |
ENST00000426156.1
ENST00000359943.3 ENST00000412653.1 |
FAM3C
|
family with sequence similarity 3, member C |
| chr19_+_11350278 | 0.63 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr14_-_100070363 | 0.63 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr20_+_44519948 | 0.63 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr12_+_50355647 | 0.63 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr3_-_57583052 | 0.63 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr11_+_45944190 | 0.62 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr7_-_98030360 | 0.62 |
ENST00000005260.8
|
BAIAP2L1
|
BAI1-associated protein 2-like 1 |
| chr19_+_51774520 | 0.62 |
ENST00000595566.1
ENST00000594261.1 |
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr2_+_207024306 | 0.61 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_-_97416400 | 0.60 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr22_+_39916558 | 0.60 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr10_+_103892787 | 0.60 |
ENST00000278070.2
ENST00000413464.2 |
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chrX_-_48326764 | 0.60 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr3_-_57583130 | 0.60 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chrX_+_131157290 | 0.59 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr20_+_44520009 | 0.58 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chr11_-_118972575 | 0.58 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr9_+_117350009 | 0.56 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chr15_-_79383102 | 0.56 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr19_-_39805976 | 0.56 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr12_+_121416437 | 0.55 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr17_-_74722536 | 0.55 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chrX_-_48326683 | 0.55 |
ENST00000440085.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chrX_+_131157322 | 0.54 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr2_+_46926048 | 0.54 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr20_-_17662878 | 0.54 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chrX_+_55246771 | 0.54 |
ENST00000289619.5
ENST00000374955.3 |
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chrX_+_153686614 | 0.53 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr2_-_65357225 | 0.53 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr10_-_127464390 | 0.53 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chrX_+_55246818 | 0.53 |
ENST00000374952.1
|
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr5_+_133984462 | 0.52 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr11_-_57282349 | 0.51 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr3_+_51428704 | 0.51 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr7_-_6523688 | 0.50 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr7_+_16793160 | 0.49 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr19_-_40730820 | 0.49 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr2_+_113403434 | 0.48 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr20_-_44519839 | 0.46 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr20_-_2821756 | 0.46 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr2_+_241526126 | 0.46 |
ENST00000391984.2
ENST00000391982.2 ENST00000404753.3 ENST00000270364.7 ENST00000352879.4 ENST00000354082.4 |
CAPN10
|
calpain 10 |
| chr17_+_39969183 | 0.45 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr1_-_111746966 | 0.45 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr8_-_143696833 | 0.45 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr11_+_32112431 | 0.45 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr17_+_74722912 | 0.44 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr5_+_68710906 | 0.43 |
ENST00000325631.5
ENST00000454295.2 |
MARVELD2
|
MARVEL domain containing 2 |
| chr19_-_13030071 | 0.43 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr6_+_32811861 | 0.42 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr6_-_28891709 | 0.42 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr5_-_158636512 | 0.42 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr8_-_144679264 | 0.41 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr22_-_35627045 | 0.41 |
ENST00000423311.1
|
CTA-714B7.5
|
CTA-714B7.5 |
| chr15_-_52030293 | 0.41 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr7_-_8301869 | 0.40 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_+_167684651 | 0.40 |
ENST00000503433.1
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr4_+_186347388 | 0.40 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr1_-_1167411 | 0.39 |
ENST00000263741.7
|
SDF4
|
stromal cell derived factor 4 |
| chr1_-_11866034 | 0.39 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr3_-_155572164 | 0.38 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_+_68711023 | 0.38 |
ENST00000515844.1
|
MARVELD2
|
MARVEL domain containing 2 |
| chr22_-_18257178 | 0.38 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr10_+_131265443 | 0.37 |
ENST00000306010.7
|
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chr21_-_28215332 | 0.37 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr2_+_201676908 | 0.37 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr11_-_777467 | 0.37 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chr5_-_176730676 | 0.37 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr11_+_67159416 | 0.36 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr1_+_180123969 | 0.36 |
ENST00000367602.3
ENST00000367600.5 |
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_-_136242956 | 0.36 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr7_-_8302164 | 0.35 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr21_+_37442239 | 0.35 |
ENST00000530908.1
ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr7_+_23749945 | 0.35 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr9_+_130565147 | 0.34 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr11_+_67776012 | 0.33 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr6_+_32811885 | 0.33 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_+_152870215 | 0.32 |
ENST00000518142.1
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr19_-_44143939 | 0.32 |
ENST00000222374.2
|
CADM4
|
cell adhesion molecule 4 |
| chr11_-_100999775 | 0.32 |
ENST00000263463.5
|
PGR
|
progesterone receptor |
| chr16_-_28074822 | 0.32 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr15_-_40213080 | 0.32 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr16_+_2533020 | 0.32 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr11_-_111957451 | 0.32 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr2_+_201676256 | 0.31 |
ENST00000452206.1
ENST00000410110.2 ENST00000409600.1 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr4_+_119200215 | 0.31 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr1_+_110162448 | 0.30 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr12_+_57623869 | 0.30 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_8301682 | 0.30 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr10_+_6392278 | 0.29 |
ENST00000391437.1
|
DKFZP667F0711
|
DKFZP667F0711 |
| chr19_-_10341948 | 0.29 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr10_-_49459800 | 0.29 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr11_-_61560053 | 0.28 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr1_-_45956822 | 0.28 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr12_+_57623477 | 0.28 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_228327943 | 0.28 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr6_-_144329384 | 0.28 |
ENST00000417959.2
|
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr2_-_27341966 | 0.28 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr3_+_51741072 | 0.25 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.7 | 2.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.7 | 3.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 1.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 2.5 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.4 | 1.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.3 | 1.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 3.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 0.8 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.3 | 2.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.7 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.9 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 2.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 2.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 2.7 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 1.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.5 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 2.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.5 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 7.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 2.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.6 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.3 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 1.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.7 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.3 | GO:1903142 | C-5 methylation of cytosine(GO:0090116) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 1.9 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:1900016 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 2.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.4 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 1.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 1.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 1.0 | GO:0006970 | response to osmotic stress(GO:0006970) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0071748 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.5 | 2.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 2.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 3.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.0 | 5.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0005655 | ribonuclease MRP complex(GO:0000172) nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.0 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.2 | 1.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.8 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 1.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 2.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 0.5 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.6 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 2.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.2 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.9 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 3.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 4.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 4.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |