Project
Illumina Body Map 2
Navigation
Downloads
Results for CTCF_CTCFL
Z-value: 1.08
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
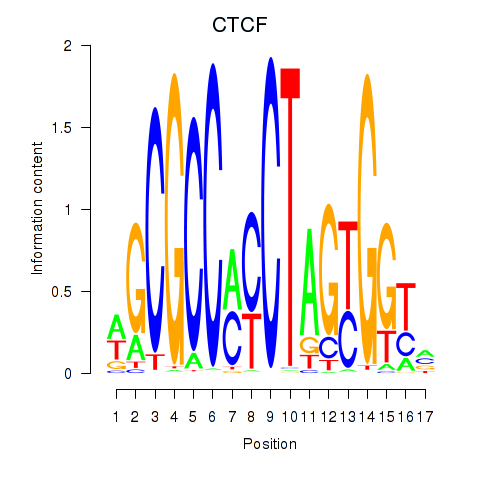
CTCF
|
ENSG00000102974.10 | CCCTC-binding factor |
|
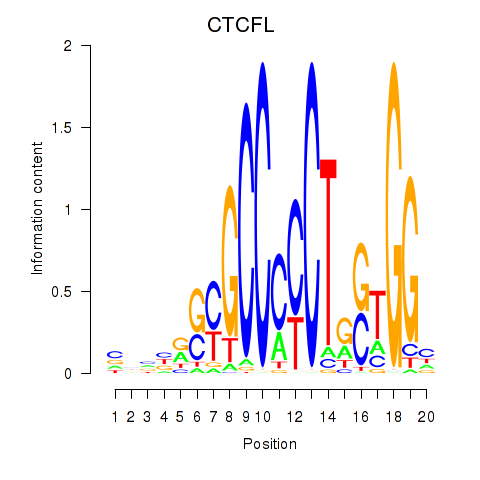
CTCFL
|
ENSG00000124092.8 | CCCTC-binding factor like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCF | hg19_v2_chr16_+_67596310_67596353 | 0.58 | 4.8e-04 | Click! |
| CTCFL | hg19_v2_chr20_-_56100155_56100163 | 0.52 | 2.3e-03 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114737146 | 5.55 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr2_+_114737472 | 3.29 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr1_-_109655377 | 3.05 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_-_109655355 | 2.86 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr11_+_61520075 | 2.85 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr15_+_45722727 | 2.66 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr4_-_44450814 | 2.57 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr1_-_156390128 | 2.46 |
ENST00000368242.3
|
C1orf61
|
chromosome 1 open reading frame 61 |
| chr2_-_154335300 | 2.43 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr11_-_64490634 | 2.37 |
ENST00000377559.3
ENST00000265459.6 |
NRXN2
|
neurexin 2 |
| chr22_-_22901636 | 2.31 |
ENST00000406503.1
ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr1_+_248100459 | 2.27 |
ENST00000366478.2
|
OR2L13
|
olfactory receptor, family 2, subfamily L, member 13 |
| chr8_+_24772455 | 2.23 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr17_+_74864476 | 2.21 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr19_+_12949251 | 2.19 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr20_+_30028322 | 2.13 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr9_+_127615733 | 2.10 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr15_-_44487408 | 2.09 |
ENST00000402883.1
ENST00000417257.1 |
FRMD5
|
FERM domain containing 5 |
| chr3_-_167098059 | 2.08 |
ENST00000392764.1
ENST00000474464.1 ENST00000392766.2 ENST00000485651.1 |
ZBBX
|
zinc finger, B-box domain containing |
| chr22_-_22901477 | 2.08 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr9_-_94712434 | 2.07 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr1_+_156890418 | 2.07 |
ENST00000337428.7
|
LRRC71
|
leucine rich repeat containing 71 |
| chr4_+_81256871 | 2.03 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr2_+_166095898 | 1.92 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_-_193059634 | 1.91 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr21_+_41239243 | 1.83 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr18_-_35065710 | 1.83 |
ENST00000589229.1
ENST00000587819.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr15_-_38856836 | 1.74 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr1_+_50513686 | 1.70 |
ENST00000448907.2
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_302918 | 1.70 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr8_-_145911188 | 1.69 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr1_+_18081804 | 1.67 |
ENST00000375406.1
|
ACTL8
|
actin-like 8 |
| chr4_-_7044657 | 1.64 |
ENST00000310085.4
|
CCDC96
|
coiled-coil domain containing 96 |
| chr5_-_179636153 | 1.63 |
ENST00000361132.4
|
RASGEF1C
|
RasGEF domain family, member 1C |
| chr2_+_186603545 | 1.61 |
ENST00000424728.1
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr16_+_67679069 | 1.55 |
ENST00000545661.1
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chrX_+_153029633 | 1.54 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr5_+_321810 | 1.48 |
ENST00000514523.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chrX_+_107020963 | 1.46 |
ENST00000509000.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr16_+_67360712 | 1.44 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr7_-_100171270 | 1.43 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr1_-_6240183 | 1.41 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr3_-_118864893 | 1.40 |
ENST00000354673.2
ENST00000425327.2 |
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr12_-_113574028 | 1.40 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr17_+_16593539 | 1.39 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr4_-_175041663 | 1.39 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr2_+_186603355 | 1.39 |
ENST00000343098.5
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr8_-_11873043 | 1.38 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr3_+_164924716 | 1.38 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr16_+_67313412 | 1.36 |
ENST00000379344.3
ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr22_-_50765489 | 1.36 |
ENST00000413817.3
|
DENND6B
|
DENN/MADD domain containing 6B |
| chr17_-_42441204 | 1.36 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr5_+_175299743 | 1.35 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr19_-_48823332 | 1.35 |
ENST00000315396.7
|
CCDC114
|
coiled-coil domain containing 114 |
| chr11_+_73675873 | 1.33 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr17_+_63133587 | 1.32 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr11_+_120382417 | 1.30 |
ENST00000527524.2
ENST00000375081.2 |
GRIK4
AP002348.1
|
glutamate receptor, ionotropic, kainate 4 Uncharacterized protein |
| chr7_-_91808441 | 1.27 |
ENST00000437357.1
ENST00000458448.1 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr16_+_67678818 | 1.27 |
ENST00000334583.6
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chr5_+_35618058 | 1.27 |
ENST00000440995.2
|
SPEF2
|
sperm flagellar 2 |
| chr5_+_35617940 | 1.25 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr4_+_89378261 | 1.25 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr3_-_108836945 | 1.24 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr10_+_6622345 | 1.23 |
ENST00000445427.1
ENST00000455810.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr3_-_108836977 | 1.23 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr10_-_49732281 | 1.22 |
ENST00000374170.1
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr4_+_7194247 | 1.20 |
ENST00000507866.2
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr12_+_51818555 | 1.20 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr22_+_19705928 | 1.18 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chrX_-_30326445 | 1.18 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr20_+_49411523 | 1.18 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr9_+_130965677 | 1.18 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chrX_-_134936735 | 1.18 |
ENST00000487941.2
ENST00000434966.2 |
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr5_+_157170703 | 1.17 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr3_+_140950612 | 1.17 |
ENST00000286353.4
ENST00000502783.1 ENST00000393010.2 ENST00000514680.1 |
ACPL2
|
acid phosphatase-like 2 |
| chr4_+_4387983 | 1.17 |
ENST00000397958.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr5_-_179050066 | 1.16 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chrX_+_149009941 | 1.15 |
ENST00000535454.1
ENST00000542674.1 ENST00000286482.1 |
MAGEA8
|
melanoma antigen family A, 8 |
| chr16_+_84002234 | 1.14 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr12_-_113573495 | 1.13 |
ENST00000446861.3
|
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr5_-_137475071 | 1.12 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr4_+_4388245 | 1.11 |
ENST00000433139.2
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr9_+_130965651 | 1.11 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr5_-_146833803 | 1.11 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr20_+_36531499 | 1.11 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr14_-_77608121 | 1.11 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger, DHHC-type containing 22 |
| chr1_+_113009163 | 1.10 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr19_+_3224700 | 1.09 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr1_+_92495528 | 1.08 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr16_-_67360662 | 1.06 |
ENST00000304372.5
|
KCTD19
|
potassium channel tetramerization domain containing 19 |
| chr19_-_11591848 | 1.06 |
ENST00000359227.3
|
ELAVL3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_+_63133526 | 1.05 |
ENST00000443584.3
|
RGS9
|
regulator of G-protein signaling 9 |
| chr11_+_66025938 | 1.05 |
ENST00000394066.2
|
KLC2
|
kinesin light chain 2 |
| chr12_+_49208234 | 1.04 |
ENST00000540990.1
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr12_+_51985001 | 1.04 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chrX_+_152783131 | 1.03 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chrX_-_134953994 | 1.03 |
ENST00000420087.2
ENST00000463085.2 ENST00000370724.3 |
CT45A4
CT45A5
|
cancer/testis antigen family 45, member A4 cancer/testis antigen family 45, member A5 |
| chr1_+_10490127 | 1.03 |
ENST00000602787.1
ENST00000602296.1 ENST00000400900.2 |
APITD1
APITD1-CORT
|
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr19_+_54926601 | 1.02 |
ENST00000301194.4
|
TTYH1
|
tweety family member 1 |
| chrX_+_134866214 | 1.02 |
ENST00000370736.3
ENST00000471213.2 |
CT45A2
|
cancer/testis antigen family 45, member A2 |
| chr1_-_156675535 | 1.01 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr16_+_71392616 | 1.01 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chrX_-_134936550 | 1.01 |
ENST00000494421.1
|
CT45A4
|
cancer/testis antigen family 45, member A4 |
| chr5_-_11904100 | 1.01 |
ENST00000359640.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr3_+_28390637 | 1.00 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr16_+_84178850 | 1.00 |
ENST00000334315.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr4_+_152198323 | 0.99 |
ENST00000455694.2
ENST00000441586.2 |
PRSS48
|
protease, serine, 48 |
| chr18_+_580367 | 0.98 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr1_-_151688528 | 0.98 |
ENST00000290585.4
|
CELF3
|
CUGBP, Elav-like family member 3 |
| chrX_-_118284542 | 0.98 |
ENST00000402510.2
|
KIAA1210
|
KIAA1210 |
| chr6_+_87646995 | 0.98 |
ENST00000305344.5
|
HTR1E
|
5-hydroxytryptamine (serotonin) receptor 1E, G protein-coupled |
| chr6_-_30043539 | 0.98 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr1_-_10003372 | 0.98 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr4_+_154265784 | 0.96 |
ENST00000240488.3
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr3_-_46759314 | 0.96 |
ENST00000315170.7
|
PRSS50
|
protease, serine, 50 |
| chr9_+_100174344 | 0.95 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr16_+_84178874 | 0.95 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr14_+_101293687 | 0.94 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_+_60691924 | 0.94 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr11_-_32452357 | 0.92 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chrX_-_154493791 | 0.92 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr3_-_48598547 | 0.92 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chrX_+_134847370 | 0.92 |
ENST00000482795.1
|
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr12_+_82752647 | 0.92 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr6_-_41747595 | 0.91 |
ENST00000373018.3
|
FRS3
|
fibroblast growth factor receptor substrate 3 |
| chr17_-_16395455 | 0.91 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr5_-_11904152 | 0.91 |
ENST00000304623.8
ENST00000458100.2 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr12_-_15374343 | 0.91 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr18_+_12093838 | 0.90 |
ENST00000587848.2
|
ANKRD62
|
ankyrin repeat domain 62 |
| chr14_+_22337014 | 0.90 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr10_+_101089107 | 0.90 |
ENST00000446890.1
ENST00000370528.3 |
CNNM1
|
cyclin M1 |
| chr9_+_100174232 | 0.90 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr1_-_111174054 | 0.90 |
ENST00000369770.3
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr9_+_138594018 | 0.89 |
ENST00000487664.1
ENST00000298480.5 ENST00000371757.2 |
KCNT1
|
potassium channel, subfamily T, member 1 |
| chr17_-_16395328 | 0.89 |
ENST00000470794.1
|
FAM211A
|
family with sequence similarity 211, member A |
| chr15_-_65715401 | 0.89 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr3_-_118864861 | 0.89 |
ENST00000441144.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr3_-_138665969 | 0.89 |
ENST00000330315.3
|
FOXL2
|
forkhead box L2 |
| chrX_+_150869023 | 0.88 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr3_-_28390415 | 0.88 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chrX_+_134883488 | 0.87 |
ENST00000370734.3
ENST00000485366.2 |
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr5_+_140186647 | 0.87 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr13_+_88324870 | 0.87 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr16_-_15950868 | 0.86 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr10_+_101088836 | 0.86 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr19_+_35634146 | 0.86 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr10_+_116853091 | 0.85 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chrX_+_103810874 | 0.85 |
ENST00000372582.1
|
IL1RAPL2
|
interleukin 1 receptor accessory protein-like 2 |
| chr19_-_49149553 | 0.85 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chrX_+_134847185 | 0.85 |
ENST00000370741.3
ENST00000497301.2 |
CT45A1
|
cancer/testis antigen family 45, member A1 |
| chr12_+_13197218 | 0.85 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr10_+_116853201 | 0.83 |
ENST00000527407.1
|
ATRNL1
|
attractin-like 1 |
| chr5_+_173416162 | 0.83 |
ENST00000340147.6
|
C5orf47
|
chromosome 5 open reading frame 47 |
| chr12_+_50451462 | 0.82 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr15_+_74165945 | 0.82 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr12_-_95510743 | 0.81 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr10_-_6622201 | 0.81 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr19_+_54926621 | 0.81 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr1_+_112298514 | 0.80 |
ENST00000536167.1
|
DDX20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr9_+_131182697 | 0.80 |
ENST00000372838.4
ENST00000411852.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr12_-_15374328 | 0.80 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr7_-_25219897 | 0.80 |
ENST00000283905.3
ENST00000409280.1 ENST00000415598.1 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr9_-_133814455 | 0.79 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr6_+_31105426 | 0.79 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr10_-_79789291 | 0.78 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr14_-_103987679 | 0.78 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr14_+_93813510 | 0.78 |
ENST00000342144.2
|
COX8C
|
cytochrome c oxidase subunit VIIIC |
| chrX_+_49593853 | 0.77 |
ENST00000376141.1
|
PAGE4
|
P antigen family, member 4 (prostate associated) |
| chr6_+_84222194 | 0.77 |
ENST00000536636.1
|
PRSS35
|
protease, serine, 35 |
| chr6_+_3849584 | 0.77 |
ENST00000380274.1
ENST00000380272.3 |
FAM50B
|
family with sequence similarity 50, member B |
| chr10_-_6622258 | 0.76 |
ENST00000263125.5
|
PRKCQ
|
protein kinase C, theta |
| chr12_+_49932886 | 0.76 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr16_-_49891694 | 0.76 |
ENST00000562520.1
|
ZNF423
|
zinc finger protein 423 |
| chr7_+_73082152 | 0.76 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr5_+_140201183 | 0.75 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr6_-_110679475 | 0.75 |
ENST00000338882.4
|
METTL24
|
methyltransferase like 24 |
| chr19_-_38747172 | 0.74 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr2_+_26395939 | 0.74 |
ENST00000401533.2
|
GAREML
|
GRB2 associated, regulator of MAPK1-like |
| chr16_-_90085824 | 0.74 |
ENST00000002501.6
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr16_+_67282853 | 0.73 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr20_-_43438912 | 0.72 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr12_-_65515334 | 0.72 |
ENST00000286574.4
|
WIF1
|
WNT inhibitory factor 1 |
| chr2_-_232791038 | 0.72 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chrX_+_114423963 | 0.72 |
ENST00000424776.3
|
RBMXL3
|
RNA binding motif protein, X-linked-like 3 |
| chr16_-_19897455 | 0.72 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr1_-_156675564 | 0.71 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr9_-_130742792 | 0.70 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr17_-_72919317 | 0.70 |
ENST00000319642.1
|
USH1G
|
Usher syndrome 1G (autosomal recessive) |
| chrX_+_49593888 | 0.69 |
ENST00000218068.6
|
PAGE4
|
P antigen family, member 4 (prostate associated) |
| chr2_-_132559234 | 0.69 |
ENST00000303798.2
|
C2orf27B
|
chromosome 2 open reading frame 27B |
| chr14_+_22217447 | 0.69 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr14_-_50319482 | 0.69 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr2_-_198650961 | 0.68 |
ENST00000282278.8
|
BOLL
|
boule-like RNA-binding protein |
| chr11_+_117103441 | 0.68 |
ENST00000531287.1
ENST00000531452.1 |
RNF214
|
ring finger protein 214 |
| chr4_-_140201333 | 0.68 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr12_-_49393092 | 0.68 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr15_+_68871308 | 0.68 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr17_+_30593195 | 0.67 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr3_+_50712672 | 0.67 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr22_-_21905750 | 0.67 |
ENST00000433039.1
|
RIMBP3C
|
RIMS binding protein 3C |
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.6 | 1.9 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.5 | 1.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 2.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 1.6 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.4 | 1.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.4 | 2.5 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.4 | 2.5 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.3 | 1.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.3 | 0.8 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 0.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.3 | 1.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 2.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 1.7 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 4.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 2.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 2.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 0.9 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 1.8 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 1.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 1.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 1.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.6 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 2.8 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.2 | 0.9 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.9 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 1.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 2.6 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 0.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.0 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.1 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.5 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 | 0.6 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 1.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) neurofibrillary tangle assembly(GO:1902988) |
| 0.1 | 1.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 1.8 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 1.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 0.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.3 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.1 | 0.1 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.3 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 2.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.7 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.7 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.4 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.1 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.6 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 1.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.5 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.1 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.3 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.6 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.5 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 1.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.2 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:1901858 | regulation of mitochondrial DNA metabolic process(GO:1901858) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 1.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 1.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.2 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 1.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 1.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 3.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.6 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 1.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0042407 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.5 | GO:0031960 | response to corticosteroid(GO:0031960) |
| 0.0 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.9 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.8 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 2.1 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 2.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.8 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 1.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 1.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.4 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0022027 | positive regulation of centrosome duplication(GO:0010825) interkinetic nuclear migration(GO:0022027) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.9 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.0 | 0.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0006325 | chromatin organization(GO:0006325) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:1901419 | regulation of vitamin D receptor signaling pathway(GO:0070562) regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 1.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0050954 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.5 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0032525 | regulation of somitogenesis(GO:0014807) somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 2.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.3 | 2.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 1.5 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 1.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 4.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 3.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 1.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.5 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.3 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 3.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 3.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 2.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.4 | 1.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 1.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.3 | 2.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 1.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.3 | 2.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 1.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.2 | 1.8 | GO:0042835 | BRE binding(GO:0042835) |
| 0.2 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 0.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 0.9 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 0.9 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 0.5 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.2 | 0.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 5.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.7 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.9 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.7 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 4.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 4.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.4 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 2.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 1.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 2.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 1.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 2.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 3.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 3.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |