Project
Illumina Body Map 2
Navigation
Downloads
Results for CUX2
Z-value: 1.50
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.9 | cut like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg19_v2_chr12_+_111537227_111537250 | 0.71 | 4.9e-06 | Click! |
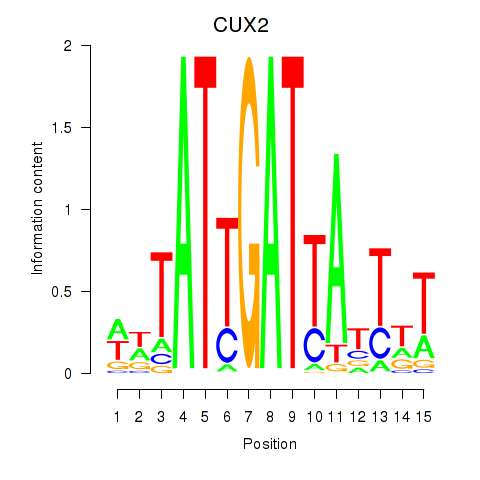
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_196912902 | 5.31 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr17_-_64216748 | 4.33 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_-_75100539 | 4.08 |
ENST00000420661.2
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr19_-_58864848 | 3.87 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr3_+_35683651 | 3.84 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr18_+_29171689 | 3.75 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr3_+_35721106 | 3.58 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_74275057 | 3.57 |
ENST00000511370.1
|
ALB
|
albumin |
| chr13_-_46679185 | 3.44 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_-_159684371 | 3.31 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr3_+_52811596 | 3.17 |
ENST00000542827.1
ENST00000273283.2 |
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr13_-_46679144 | 3.06 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr4_+_69681710 | 3.06 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr3_+_35682913 | 3.04 |
ENST00000449196.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_35683775 | 2.98 |
ENST00000452563.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr14_-_21056121 | 2.83 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr5_-_135290705 | 2.81 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_+_57320437 | 2.81 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr12_-_78753496 | 2.81 |
ENST00000548512.1
|
RP11-38F22.1
|
RP11-38F22.1 |
| chr12_+_111471828 | 2.76 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr3_+_35683816 | 2.68 |
ENST00000438577.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_211342432 | 2.59 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr14_+_88490894 | 2.57 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr4_-_72649763 | 2.50 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr17_-_64225508 | 2.46 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_187187098 | 2.42 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr5_+_173472607 | 2.31 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr8_+_77593448 | 2.26 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_+_161190160 | 2.23 |
ENST00000594609.1
|
AL590714.1
|
Uncharacterized protein |
| chr3_+_35721182 | 2.16 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_35440796 | 2.09 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_96829246 | 2.06 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr10_+_95517660 | 2.06 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr10_+_95517616 | 2.05 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr19_-_16008880 | 2.04 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr7_+_151653464 | 2.00 |
ENST00000431418.2
ENST00000392800.2 |
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr3_-_10452359 | 1.97 |
ENST00000452124.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr8_+_77593474 | 1.96 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr11_-_35440579 | 1.95 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr6_-_49931818 | 1.91 |
ENST00000322066.3
|
DEFB114
|
defensin, beta 114 |
| chr10_+_95517566 | 1.86 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr19_+_45409011 | 1.81 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr3_-_148939598 | 1.76 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr13_-_62001982 | 1.71 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr20_-_22566089 | 1.70 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr15_+_51973680 | 1.66 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr16_-_48281305 | 1.64 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr2_+_108863651 | 1.64 |
ENST00000329106.2
ENST00000376700.1 |
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3 |
| chr3_-_108836977 | 1.64 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr22_-_50523843 | 1.63 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr19_+_3224700 | 1.59 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr3_-_42452050 | 1.59 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chr11_-_49230184 | 1.58 |
ENST00000340334.7
ENST00000256999.2 |
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr12_-_10151773 | 1.57 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr11_+_58706944 | 1.56 |
ENST00000532726.1
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr1_+_50571949 | 1.54 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_22601 | 1.51 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr11_-_49230144 | 1.47 |
ENST00000343844.4
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr12_+_20968608 | 1.46 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr4_+_187187337 | 1.45 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chrX_+_55246818 | 1.45 |
ENST00000374952.1
|
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr1_+_196857144 | 1.45 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr5_-_135290651 | 1.45 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr9_-_27297126 | 1.43 |
ENST00000380031.1
ENST00000537675.1 ENST00000380032.3 |
EQTN
|
equatorin, sperm acrosome associated |
| chr1_-_169733789 | 1.42 |
ENST00000454271.1
ENST00000609271.1 |
RP1-117P20.3
SELE
|
RP1-117P20.3 selectin E |
| chr3_+_53528659 | 1.41 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr3_+_35721130 | 1.41 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_+_152850787 | 1.41 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr6_+_73076432 | 1.40 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr8_-_86290333 | 1.38 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr3_+_63428752 | 1.37 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr19_-_36297632 | 1.36 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_-_197036364 | 1.36 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr4_-_139051839 | 1.36 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr4_-_34271356 | 1.35 |
ENST00000514877.1
|
RP11-548L20.1
|
RP11-548L20.1 |
| chr15_+_51973708 | 1.34 |
ENST00000558709.1
|
SCG3
|
secretogranin III |
| chr15_-_79383102 | 1.33 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr4_-_176733377 | 1.33 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr7_-_121944491 | 1.32 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr3_+_181429704 | 1.31 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr22_+_51176624 | 1.29 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr3_+_134514093 | 1.29 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr2_+_234959323 | 1.29 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr12_-_133186983 | 1.28 |
ENST00000544018.2
|
LRCOL1
|
leucine rich colipase-like 1 |
| chr1_-_23340400 | 1.25 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr1_-_68915610 | 1.25 |
ENST00000262340.5
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa |
| chr9_+_74729511 | 1.24 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr2_-_220174166 | 1.24 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr5_-_146302078 | 1.23 |
ENST00000508545.2
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_106161540 | 1.23 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr1_+_207262627 | 1.23 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr12_-_45269430 | 1.23 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr11_-_89653576 | 1.22 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr6_-_154677866 | 1.22 |
ENST00000367220.4
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr15_+_54901540 | 1.22 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr5_-_110062384 | 1.22 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr3_-_172859017 | 1.22 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr22_-_50523760 | 1.21 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr20_-_52612705 | 1.21 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr11_-_57158109 | 1.20 |
ENST00000525955.1
ENST00000533605.1 ENST00000311862.5 |
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chr11_-_110561721 | 1.19 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr8_-_86253888 | 1.19 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr2_+_211421262 | 1.19 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr17_+_43922241 | 1.18 |
ENST00000329196.5
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr6_-_49834240 | 1.18 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chrX_+_55246771 | 1.18 |
ENST00000289619.5
ENST00000374955.3 |
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr21_-_22175341 | 1.17 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_-_76398077 | 1.16 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr6_-_49834209 | 1.16 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr8_+_104892639 | 1.16 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr13_+_78315528 | 1.16 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_-_38008783 | 1.15 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr12_+_75728419 | 1.13 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr3_+_26735991 | 1.13 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr6_-_154677900 | 1.12 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_+_51549498 | 1.12 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_+_240255166 | 1.11 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr3_-_165555200 | 1.10 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_-_55412774 | 1.09 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr10_+_5238793 | 1.09 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr7_-_96132835 | 1.07 |
ENST00000356686.1
|
C7orf76
|
chromosome 7 open reading frame 76 |
| chr22_-_50523807 | 1.07 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chrX_-_80457385 | 1.07 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr14_-_47812321 | 1.07 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr11_-_49230084 | 1.05 |
ENST00000356696.3
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr4_+_71263599 | 1.05 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr17_+_56315787 | 1.05 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr12_-_45270151 | 1.04 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr12_-_45270077 | 1.03 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr2_-_152830479 | 1.03 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_-_110062349 | 1.03 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr20_-_52612468 | 1.02 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr2_+_102953608 | 1.02 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr12_-_7818474 | 1.02 |
ENST00000229304.4
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr12_-_55375622 | 1.01 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr9_-_21187598 | 1.01 |
ENST00000421715.1
|
IFNA4
|
interferon, alpha 4 |
| chrX_-_9734004 | 1.01 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr18_+_50278430 | 1.01 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr8_+_36641842 | 0.99 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr14_-_21055874 | 0.99 |
ENST00000553849.1
|
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr9_+_105757590 | 0.99 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_+_51863433 | 0.99 |
ENST00000444293.1
|
IQCF3
|
IQ motif containing F3 |
| chr11_-_62997124 | 0.98 |
ENST00000306494.6
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr2_+_234959376 | 0.98 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr10_+_124670121 | 0.98 |
ENST00000368894.1
|
FAM24A
|
family with sequence similarity 24, member A |
| chr7_-_14942283 | 0.98 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr6_+_127898312 | 0.97 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr10_+_68685764 | 0.97 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_100354442 | 0.96 |
ENST00000475887.1
|
GPR128
|
G protein-coupled receptor 128 |
| chr22_-_50523688 | 0.96 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_45269769 | 0.96 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr3_-_108836945 | 0.95 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr8_-_120651020 | 0.94 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr7_-_88425025 | 0.94 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr16_+_58283814 | 0.94 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr15_+_42651691 | 0.94 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr4_+_69962212 | 0.94 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_-_19615744 | 0.93 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr12_-_45269251 | 0.92 |
ENST00000553120.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr4_-_70505358 | 0.92 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr7_-_18067478 | 0.92 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr4_-_34041504 | 0.92 |
ENST00000512581.1
ENST00000505018.1 |
RP11-79E3.3
|
RP11-79E3.3 |
| chr19_-_3786354 | 0.91 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr4_+_159442878 | 0.91 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr19_+_54466179 | 0.91 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr5_+_121465207 | 0.90 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr5_-_140683612 | 0.90 |
ENST00000239451.4
|
SLC25A2
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 2 |
| chr11_+_60467047 | 0.90 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr14_-_74892805 | 0.90 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr4_-_110736505 | 0.89 |
ENST00000609440.1
|
RP11-602N24.3
|
RP11-602N24.3 |
| chr1_+_207262578 | 0.89 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr1_+_28099683 | 0.89 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr7_-_14942467 | 0.88 |
ENST00000407950.1
ENST00000444700.2 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_81763184 | 0.87 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_+_49178536 | 0.87 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr6_+_35748783 | 0.87 |
ENST00000373861.5
ENST00000542261.1 |
CLPSL1
|
colipase-like 1 |
| chrX_+_30261847 | 0.87 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr4_+_155484155 | 0.86 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_71587669 | 0.86 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_-_47736896 | 0.85 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr12_-_64784306 | 0.84 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr15_+_42697065 | 0.84 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr10_+_124320156 | 0.84 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_81763127 | 0.83 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr22_+_18721427 | 0.83 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr4_+_159443090 | 0.82 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr19_+_18726786 | 0.82 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr10_-_79398250 | 0.82 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_69962185 | 0.81 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr22_+_41074754 | 0.81 |
ENST00000249016.4
|
MCHR1
|
melanin-concentrating hormone receptor 1 |
| chr4_+_74347400 | 0.81 |
ENST00000226355.3
|
AFM
|
afamin |
| chr10_-_28270795 | 0.80 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr11_+_125658006 | 0.80 |
ENST00000445202.1
|
PATE3
|
prostate and testis expressed 3 |
| chr4_-_150736962 | 0.80 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chrX_+_49363665 | 0.79 |
ENST00000381700.6
|
GAGE1
|
G antigen 1 |
| chr11_+_134144139 | 0.79 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr1_+_207262540 | 0.79 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr2_-_152830441 | 0.79 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_+_128349106 | 0.79 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr1_+_66820058 | 0.78 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.3 | 3.8 | GO:1903718 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 1.2 | 3.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 3.9 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.7 | 2.0 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.6 | 6.8 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.6 | 1.8 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) positive regulation of receptor catabolic process(GO:2000646) |
| 0.5 | 2.0 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.4 | 1.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.4 | 1.7 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.4 | 1.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.4 | 4.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 5.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 1.1 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.4 | 1.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.4 | 2.6 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.4 | 1.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 2.5 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 1.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.3 | 2.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 3.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 3.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 1.9 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.3 | 1.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 1.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.3 | 1.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 5.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 0.8 | GO:2001202 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.3 | 3.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 1.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 2.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.8 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.3 | 1.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 | 1.3 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.2 | 4.4 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 1.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 3.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 1.0 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 1.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 3.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 2.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.9 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 3.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 2.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 2.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.3 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.1 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.4 | GO:0001808 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 2.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 1.5 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 1.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 3.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 17.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 1.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.7 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.4 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.1 | 2.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 3.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.8 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 3.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 3.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 2.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 2.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 2.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.0 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 5.4 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.1 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 1.0 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:1902953 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 2.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 2.7 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.4 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.0 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 1.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 2.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.5 | 9.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 1.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.4 | 1.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.3 | 1.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.3 | 2.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.2 | 1.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 2.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 2.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 4.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 3.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 12.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 5.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 2.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 4.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 3.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.9 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 1.0 | 6.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 3.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.9 | 4.4 | GO:0033265 | choline binding(GO:0033265) |
| 0.8 | 2.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.7 | 2.0 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.7 | 2.0 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.5 | 1.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.4 | 1.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.4 | 2.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.4 | 1.6 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.4 | 3.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 1.0 | GO:0072544 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.3 | 1.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.3 | 2.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 7.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 2.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 3.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 3.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 2.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 5.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 1.9 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 1.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 3.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 2.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.8 | GO:0055103 | titin binding(GO:0031432) ligase regulator activity(GO:0055103) |
| 0.1 | 0.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 2.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 21.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 4.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 3.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 6.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 2.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 10.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 7.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 6.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 6.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 3.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 3.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 5.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |