Project
Illumina Body Map 2
Navigation
Downloads
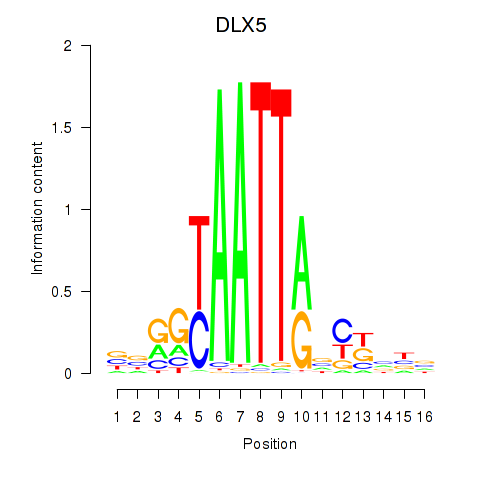
Results for DLX5
Z-value: 0.72
Transcription factors associated with DLX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX5
|
ENSG00000105880.4 | distal-less homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX5 | hg19_v2_chr7_-_96654133_96654409 | 0.58 | 4.6e-04 | Click! |
Activity profile of DLX5 motif
Sorted Z-values of DLX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_124553437 | 4.11 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr1_+_160160283 | 2.88 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr12_+_54378923 | 2.75 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr1_+_160160346 | 2.71 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr14_+_32798547 | 2.14 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_81110684 | 1.97 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr14_+_32798462 | 1.90 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_+_169575875 | 1.84 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_+_186560476 | 1.72 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr3_-_105588231 | 1.68 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr13_-_36050819 | 1.66 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr3_+_186560462 | 1.66 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr11_-_102709441 | 1.66 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr12_+_54378849 | 1.44 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr1_+_202317815 | 1.38 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chrX_+_100224676 | 1.38 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr11_-_26593779 | 1.30 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr20_-_56647116 | 1.24 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr11_-_26593677 | 1.23 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr1_+_202317855 | 1.22 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr3_+_2933893 | 1.16 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr7_+_70597109 | 1.15 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr10_-_97200772 | 1.12 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr13_-_36788718 | 1.06 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr4_-_138453559 | 1.05 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr11_-_26593649 | 1.04 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr5_-_58571935 | 0.99 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_+_818902 | 0.93 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr3_-_105587879 | 0.88 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_7013897 | 0.86 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr1_-_112903150 | 0.86 |
ENST00000427290.1
|
RP5-965F6.2
|
RP5-965F6.2 |
| chr6_-_78173490 | 0.85 |
ENST00000369947.2
|
HTR1B
|
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| chr16_-_52061283 | 0.81 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr2_+_228736321 | 0.79 |
ENST00000309931.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr13_+_110958124 | 0.76 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr4_+_175839517 | 0.74 |
ENST00000502940.1
ENST00000502305.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr8_-_7332604 | 0.74 |
ENST00000316169.2
|
DEFB104B
|
defensin, beta 104B |
| chr9_-_34329198 | 0.72 |
ENST00000379166.2
ENST00000345050.2 |
KIF24
|
kinesin family member 24 |
| chr12_+_7014064 | 0.70 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr18_-_33709268 | 0.70 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr18_-_44181442 | 0.68 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr20_+_58571419 | 0.67 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr4_-_74853897 | 0.64 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr14_-_95236551 | 0.63 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr11_-_102651343 | 0.63 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_-_143767428 | 0.62 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr2_+_228735763 | 0.60 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr17_+_43238438 | 0.60 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr17_+_36508826 | 0.60 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr7_-_99716940 | 0.59 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_-_28969517 | 0.57 |
ENST00000263974.4
ENST00000373824.4 |
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr7_-_99716952 | 0.56 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_95963052 | 0.54 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr3_+_11178779 | 0.54 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr2_+_169757750 | 0.52 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr12_+_7014126 | 0.51 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_-_95774672 | 0.50 |
ENST00000549961.1
|
RP11-167N24.6
|
Uncharacterized protein |
| chr19_+_36239576 | 0.49 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr22_-_32860427 | 0.48 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chr7_-_99717463 | 0.48 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_-_33913708 | 0.48 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr17_+_28268623 | 0.48 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_+_7456880 | 0.44 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr16_+_58010339 | 0.43 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr1_-_197115818 | 0.43 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr7_-_86849836 | 0.42 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr8_-_94179079 | 0.41 |
ENST00000521906.1
|
C8orf87
|
chromosome 8 open reading frame 87 |
| chr18_-_53255379 | 0.40 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr11_-_123814580 | 0.36 |
ENST00000321252.2
|
OR6T1
|
olfactory receptor, family 6, subfamily T, member 1 |
| chr1_-_242612779 | 0.36 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr15_+_63188009 | 0.36 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr12_+_52695617 | 0.36 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chrX_+_100645812 | 0.35 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr1_+_61547894 | 0.34 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr19_-_36606181 | 0.34 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr15_+_89164520 | 0.34 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr6_-_33256664 | 0.32 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr2_-_183387064 | 0.32 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_-_73051037 | 0.32 |
ENST00000445814.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr1_+_54569968 | 0.32 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chr17_-_38956205 | 0.31 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr17_+_59489112 | 0.31 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr8_-_10512569 | 0.31 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr19_+_58570605 | 0.31 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr15_+_41913690 | 0.30 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr3_-_52567792 | 0.30 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr10_-_128110441 | 0.29 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr4_-_66536057 | 0.26 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr4_-_8873531 | 0.26 |
ENST00000400677.3
|
HMX1
|
H6 family homeobox 1 |
| chr1_-_151762900 | 0.26 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr13_+_36050881 | 0.25 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr1_+_61547405 | 0.24 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr1_-_242612726 | 0.24 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr17_+_42264556 | 0.23 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr5_-_92023923 | 0.22 |
ENST00000503489.1
|
CTC-529L17.2
|
CTC-529L17.2 |
| chr13_+_111748183 | 0.20 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr18_+_63091237 | 0.20 |
ENST00000582028.1
|
RP11-453M23.1
|
RP11-453M23.1 |
| chr20_+_43849941 | 0.20 |
ENST00000372769.3
|
SEMG2
|
semenogelin II |
| chr2_+_228736335 | 0.19 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr12_-_10978957 | 0.18 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr1_-_78444776 | 0.16 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr3_-_180397256 | 0.16 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr1_-_150669604 | 0.15 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_+_32579321 | 0.15 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr7_-_7575477 | 0.14 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr2_+_30369807 | 0.14 |
ENST00000379520.3
ENST00000379519.3 ENST00000261353.4 |
YPEL5
|
yippee-like 5 (Drosophila) |
| chrX_-_16887963 | 0.14 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_-_183387430 | 0.14 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr19_+_6372444 | 0.14 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr5_-_160279207 | 0.14 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr8_-_82373809 | 0.13 |
ENST00000379071.2
|
FABP9
|
fatty acid binding protein 9, testis |
| chr3_+_194426277 | 0.13 |
ENST00000439479.1
|
AC106706.1
|
AC106706.1 |
| chr19_+_4007644 | 0.13 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr6_-_27860956 | 0.12 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr19_+_36606354 | 0.11 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr4_+_66536248 | 0.10 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr14_-_68283291 | 0.10 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chr4_-_66536196 | 0.10 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr8_+_32579271 | 0.10 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr18_+_55888767 | 0.10 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_30369859 | 0.09 |
ENST00000402003.3
|
YPEL5
|
yippee-like 5 (Drosophila) |
| chr11_-_26743546 | 0.08 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_-_53171128 | 0.06 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr17_-_37009882 | 0.06 |
ENST00000378096.3
ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23
|
ribosomal protein L23 |
| chr17_-_39341594 | 0.06 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr19_+_54641444 | 0.06 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr3_-_186288097 | 0.06 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr7_+_97736197 | 0.05 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr2_-_220118631 | 0.05 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr12_+_51818749 | 0.05 |
ENST00000514353.3
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr6_+_27861190 | 0.04 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr1_-_21620877 | 0.04 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_78444738 | 0.03 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.5 | 9.7 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 | 4.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.3 | 4.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.2 | 0.8 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.7 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 2.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.9 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.2 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 3.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 1.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.5 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 1.7 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 4.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 4.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 4.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 2.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 4.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.6 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.8 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 2.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 3.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |