Project
Illumina Body Map 2
Navigation
Downloads
Results for DPRX
Z-value: 0.89
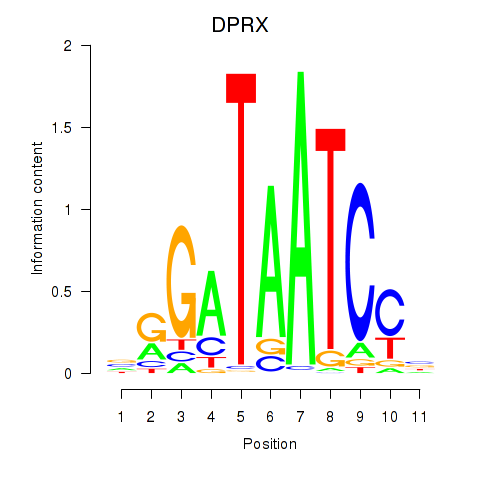
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DPRX | hg19_v2_chr19_+_54135310_54135310 | -0.33 | 6.8e-02 | Click! |
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_64271316 | 2.83 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr18_-_64271363 | 2.43 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr10_-_104178857 | 1.86 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr6_-_84418432 | 1.56 |
ENST00000519825.1
ENST00000523484.2 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr18_-_40857493 | 1.48 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr6_-_84418841 | 1.41 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr16_+_81272287 | 1.36 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr16_-_15950868 | 1.34 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chrX_+_100333709 | 1.22 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chrX_-_138724994 | 1.22 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr20_+_10199566 | 1.22 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr12_+_6949964 | 1.18 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr6_-_84418860 | 1.18 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr2_-_235405168 | 1.08 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr1_+_20396649 | 1.06 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr13_-_45775162 | 1.00 |
ENST00000405872.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr1_+_50569575 | 0.96 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chrX_-_138724677 | 0.93 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr9_-_23821273 | 0.93 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr22_+_23161491 | 0.92 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr2_-_158184211 | 0.91 |
ENST00000397283.2
|
ERMN
|
ermin, ERM-like protein |
| chr14_+_92789498 | 0.86 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr14_+_22670455 | 0.86 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr7_-_135412925 | 0.85 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr22_-_38480100 | 0.85 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr18_-_70532906 | 0.85 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr6_-_84418738 | 0.84 |
ENST00000519779.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr1_+_115642293 | 0.77 |
ENST00000448680.1
|
RP4-666F24.3
|
RP4-666F24.3 |
| chr14_-_101034407 | 0.76 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr5_-_66492562 | 0.74 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr15_+_34260921 | 0.74 |
ENST00000560035.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr14_+_62585332 | 0.72 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr20_-_54580523 | 0.72 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chr14_+_22265444 | 0.71 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr16_-_30022735 | 0.71 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr22_+_32754139 | 0.69 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr4_+_36283213 | 0.68 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr21_+_19617140 | 0.68 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr6_+_43215727 | 0.68 |
ENST00000304139.5
|
TTBK1
|
tau tubulin kinase 1 |
| chr1_+_36023370 | 0.67 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr11_+_61522844 | 0.66 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr8_+_65285851 | 0.66 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr6_-_40555176 | 0.64 |
ENST00000338305.6
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr17_+_12626199 | 0.64 |
ENST00000609971.1
|
AC005358.1
|
AC005358.1 |
| chr6_-_128239749 | 0.64 |
ENST00000537166.1
|
THEMIS
|
thymocyte selection associated |
| chr19_+_39930212 | 0.63 |
ENST00000396843.1
|
AC011500.1
|
Interleukin-like; Uncharacterized protein |
| chr6_-_128239685 | 0.63 |
ENST00000368250.1
|
THEMIS
|
thymocyte selection associated |
| chr1_-_102312600 | 0.61 |
ENST00000359814.3
|
OLFM3
|
olfactomedin 3 |
| chr8_-_79717750 | 0.60 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr18_-_3845321 | 0.57 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_-_103997862 | 0.56 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr2_-_30144432 | 0.55 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr5_-_45696253 | 0.54 |
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr20_+_123010 | 0.54 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr14_+_22204418 | 0.52 |
ENST00000390426.2
|
TRAV4
|
T cell receptor alpha variable 4 |
| chr4_-_103998060 | 0.52 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr10_+_118187379 | 0.52 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr8_-_95274536 | 0.52 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr11_-_10715287 | 0.51 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr11_-_2323290 | 0.50 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr8_-_79717163 | 0.50 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr4_-_164534657 | 0.50 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr8_-_120605194 | 0.50 |
ENST00000522167.1
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr4_+_159131630 | 0.49 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr15_+_29211570 | 0.49 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr12_-_71182695 | 0.48 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr19_-_13734804 | 0.47 |
ENST00000574974.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr17_-_72348271 | 0.46 |
ENST00000599136.1
|
AC103809.2
|
Uncharacterized protein; cDNA FLJ33592 fis, clone BRAMY2012691 |
| chr1_+_207262578 | 0.46 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_152086556 | 0.46 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr3_+_111393501 | 0.46 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr15_+_96904487 | 0.46 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr1_+_36024107 | 0.44 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr4_-_103998439 | 0.44 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr10_-_72362515 | 0.44 |
ENST00000373209.2
ENST00000441259.1 |
PRF1
|
perforin 1 (pore forming protein) |
| chr1_-_102312517 | 0.43 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr7_+_28452130 | 0.43 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr11_-_104905840 | 0.43 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_+_207262627 | 0.42 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_+_120049826 | 0.41 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr2_-_26700900 | 0.41 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr22_-_19137796 | 0.41 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr1_+_74701062 | 0.40 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_+_71996972 | 0.39 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr8_+_38244638 | 0.39 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr12_-_95010147 | 0.39 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr18_-_3845293 | 0.38 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr14_-_101034811 | 0.38 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr1_+_207262170 | 0.38 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr14_-_106518922 | 0.38 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr4_-_123542224 | 0.38 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr17_+_48911942 | 0.38 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr3_-_167371740 | 0.37 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr4_+_57396766 | 0.36 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr7_-_84121858 | 0.36 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_-_41122063 | 0.36 |
ENST00000426005.2
ENST00000437044.2 ENST00000373127.4 |
TREML1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr12_-_9913489 | 0.36 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr3_+_10068095 | 0.36 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr4_+_113970772 | 0.36 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr5_+_132083106 | 0.35 |
ENST00000378731.1
|
CCNI2
|
cyclin I family, member 2 |
| chr8_-_101157680 | 0.35 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr1_+_190448095 | 0.35 |
ENST00000424735.1
|
RP11-547I7.2
|
RP11-547I7.2 |
| chr9_-_13432977 | 0.35 |
ENST00000605459.1
|
RP11-536O18.2
|
RP11-536O18.2 |
| chr7_+_134464376 | 0.35 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr13_-_44980021 | 0.35 |
ENST00000432701.2
ENST00000607312.1 |
LINC01071
|
long intergenic non-protein coding RNA 1071 |
| chr17_+_43318434 | 0.35 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chrX_-_140336629 | 0.34 |
ENST00000358993.2
|
SPANXC
|
SPANX family, member C |
| chr17_-_19771216 | 0.34 |
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2 |
| chr20_+_1875942 | 0.34 |
ENST00000358771.4
|
SIRPA
|
signal-regulatory protein alpha |
| chr3_+_45984947 | 0.34 |
ENST00000304552.4
|
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr14_-_21056121 | 0.34 |
ENST00000557105.1
ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr1_-_173174681 | 0.33 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chrX_+_15767971 | 0.33 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr11_+_60383204 | 0.33 |
ENST00000412599.1
ENST00000320202.4 |
LINC00301
|
long intergenic non-protein coding RNA 301 |
| chr1_+_202431859 | 0.33 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_142224280 | 0.32 |
ENST00000390367.3
|
TRBV11-1
|
T cell receptor beta variable 11-1 |
| chr4_-_87374330 | 0.32 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_+_132757215 | 0.31 |
ENST00000321871.6
ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108
|
transmembrane protein 108 |
| chr18_+_32290218 | 0.31 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr17_-_15244894 | 0.31 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chrX_+_139791917 | 0.30 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr11_-_2323089 | 0.30 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr3_+_40351169 | 0.30 |
ENST00000232905.3
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr11_+_61248583 | 0.29 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr5_+_169010638 | 0.29 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr5_-_54988559 | 0.29 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr4_-_87279520 | 0.29 |
ENST00000506773.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_14628234 | 0.29 |
ENST00000595139.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr15_-_83240783 | 0.29 |
ENST00000568994.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_+_7118755 | 0.29 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr15_+_34261089 | 0.29 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr1_+_202317815 | 0.29 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_-_236046872 | 0.28 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr1_-_209741018 | 0.28 |
ENST00000424696.2
|
RP1-272L16.1
|
RP1-272L16.1 |
| chr6_-_32784687 | 0.28 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr14_-_69446034 | 0.28 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr8_-_145701718 | 0.28 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr12_-_7848364 | 0.27 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr11_-_85430356 | 0.27 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr1_+_207262540 | 0.27 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr18_+_13465008 | 0.27 |
ENST00000593236.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_-_104916034 | 0.27 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr2_+_33701286 | 0.27 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr6_+_31555045 | 0.27 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr8_+_72755367 | 0.27 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr19_-_14628645 | 0.27 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_+_2323236 | 0.27 |
ENST00000182290.4
|
TSPAN32
|
tetraspanin 32 |
| chr17_-_73150629 | 0.27 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr12_+_54891495 | 0.27 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr22_-_36903101 | 0.26 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr20_+_8112824 | 0.26 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr14_+_22931924 | 0.26 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr14_-_22005343 | 0.26 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr14_-_22005062 | 0.25 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr3_+_111393659 | 0.25 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr9_+_37667978 | 0.25 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr16_-_46723066 | 0.25 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr8_-_37411648 | 0.25 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr6_+_130339710 | 0.25 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr11_+_2323349 | 0.24 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr11_-_65629497 | 0.24 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr18_-_67614645 | 0.24 |
ENST00000577287.1
|
CD226
|
CD226 molecule |
| chr17_+_41150290 | 0.24 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr1_+_111415757 | 0.24 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr12_+_113344755 | 0.24 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr14_+_77292715 | 0.24 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr11_-_104972158 | 0.24 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr22_-_36903069 | 0.24 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr12_+_86268065 | 0.24 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr14_-_22005197 | 0.23 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr7_+_94537247 | 0.23 |
ENST00000422324.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_-_8816375 | 0.23 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr14_-_21055874 | 0.22 |
ENST00000553849.1
|
RNASE11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr17_-_48785216 | 0.22 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr9_+_17134980 | 0.22 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr16_+_63764128 | 0.22 |
ENST00000563061.1
|
RP11-370P15.2
|
RP11-370P15.2 |
| chr11_+_111385497 | 0.22 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr8_+_104384616 | 0.21 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr4_-_122791583 | 0.21 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr16_+_67918708 | 0.21 |
ENST00000339176.3
ENST00000576758.1 |
NRN1L
|
neuritin 1-like |
| chr8_+_110656344 | 0.21 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr5_-_9712312 | 0.21 |
ENST00000506620.1
ENST00000514078.1 ENST00000606744.1 |
TAS2R1
CTD-2143L24.1
|
taste receptor, type 2, member 1 CTD-2143L24.1 |
| chr13_+_52436111 | 0.21 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr4_-_87374283 | 0.21 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_-_112564797 | 0.21 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr1_-_65533390 | 0.20 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr6_+_31554826 | 0.20 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr1_+_11824462 | 0.20 |
ENST00000435090.1
|
C1orf167
|
chromosome 1 open reading frame 167 |
| chr1_-_170043709 | 0.20 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr14_-_106380248 | 0.20 |
ENST00000390590.1
|
IGHD3-3
|
immunoglobulin heavy diversity 3-3 |
| chr12_+_103545593 | 0.20 |
ENST00000547418.1
|
RP11-552I14.1
|
Uncharacterized protein |
| chr14_+_23009190 | 0.20 |
ENST00000390532.1
|
TRAJ5
|
T cell receptor alpha joining 5 |
| chr4_-_87279641 | 0.19 |
ENST00000512689.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_+_41284121 | 0.19 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr14_-_22005018 | 0.19 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_-_49523896 | 0.19 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr6_-_35480640 | 0.19 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr1_-_150780757 | 0.19 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr7_+_50135433 | 0.19 |
ENST00000297001.6
|
C7orf72
|
chromosome 7 open reading frame 72 |
| chr3_-_158390282 | 0.19 |
ENST00000264265.3
|
LXN
|
latexin |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 0.3 | 1.0 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.3 | 1.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.3 | 1.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 1.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 1.4 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 5.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.5 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.1 | GO:0002652 | regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.3 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.2 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 0.3 | GO:1903181 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.5 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.4 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.2 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.4 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.6 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.3 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 1.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 2.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 0.5 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.9 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.2 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 1.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 5.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 5.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.2 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.4 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.9 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |