Project
Illumina Body Map 2
Navigation
Downloads
Results for DUXA
Z-value: 1.33
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.2 | double homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DUXA | hg19_v2_chr19_-_57678811_57678811 | -0.05 | 7.8e-01 | Click! |
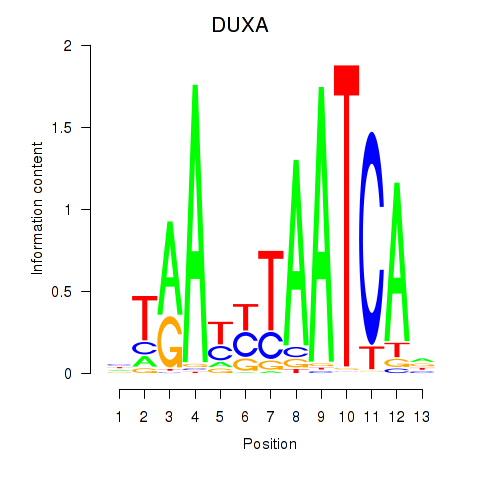
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175629164 | 7.49 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr16_+_6533729 | 5.22 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_175629135 | 3.71 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr10_-_69425399 | 3.55 |
ENST00000330298.6
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr16_+_6533380 | 3.54 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_+_173472607 | 3.53 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr11_-_16419067 | 3.53 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr4_-_46126093 | 3.38 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr10_+_69865866 | 3.24 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr7_+_54398390 | 3.23 |
ENST00000458615.1
ENST00000426205.1 |
RP11-436F9.1
|
RP11-436F9.1 |
| chr9_-_23779367 | 3.09 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr4_-_6557336 | 3.03 |
ENST00000507294.1
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr11_-_89653576 | 2.83 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr22_+_31518938 | 2.71 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_-_166930131 | 2.67 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr11_+_55029628 | 2.63 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr10_-_98118724 | 2.59 |
ENST00000393870.2
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr19_-_51472031 | 2.53 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr19_+_52932435 | 2.53 |
ENST00000301085.4
|
ZNF534
|
zinc finger protein 534 |
| chr9_-_28670283 | 2.53 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr2_+_149632783 | 2.52 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr2_-_224467093 | 2.25 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr19_-_51471381 | 2.24 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr15_+_80733570 | 2.18 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr19_-_51471362 | 2.04 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr6_+_53883708 | 1.94 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr6_+_35748783 | 1.94 |
ENST00000373861.5
ENST00000542261.1 |
CLPSL1
|
colipase-like 1 |
| chr15_-_26874230 | 1.93 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr15_+_71228826 | 1.93 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_80697396 | 1.86 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr11_-_117688216 | 1.84 |
ENST00000525836.1
|
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr7_-_14942467 | 1.81 |
ENST00000407950.1
ENST00000444700.2 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_-_181160240 | 1.81 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr9_+_139877445 | 1.79 |
ENST00000408973.2
|
LCNL1
|
lipocalin-like 1 |
| chrX_+_18725758 | 1.79 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr12_+_26164645 | 1.77 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_234667504 | 1.75 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr7_-_14942283 | 1.73 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr6_+_73076432 | 1.66 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr10_+_68685764 | 1.66 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr16_-_57219721 | 1.65 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr1_+_92632542 | 1.63 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_+_166428839 | 1.62 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr19_-_57967854 | 1.57 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr15_+_45028520 | 1.53 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_-_165414414 | 1.53 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr10_+_18689637 | 1.52 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_+_48485341 | 1.52 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr16_-_75734044 | 1.52 |
ENST00000398113.2
ENST00000538623.1 |
AC025287.1
|
Uncharacterized protein |
| chr17_-_57158523 | 1.50 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr16_+_76587314 | 1.50 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr10_-_29811456 | 1.50 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr8_-_107782463 | 1.49 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chrX_+_85969626 | 1.49 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chrX_-_72095808 | 1.47 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
| chr8_-_7673238 | 1.45 |
ENST00000335021.2
|
DEFB107A
|
defensin, beta 107A |
| chr3_+_108321623 | 1.45 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr21_+_34398153 | 1.44 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr8_+_7353368 | 1.44 |
ENST00000355602.2
|
DEFB107B
|
defensin, beta 107B |
| chr8_-_7343922 | 1.44 |
ENST00000335479.2
|
DEFB106B
|
defensin, beta 106B |
| chr12_-_81763184 | 1.42 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_+_69501943 | 1.41 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr11_+_49050504 | 1.41 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chr6_+_155538093 | 1.39 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr10_-_61900762 | 1.39 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_-_128684488 | 1.39 |
ENST00000511204.1
|
RP11-723O4.6
|
Uncharacterized protein FLJ43738 |
| chr16_-_21314360 | 1.38 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chrX_-_138724677 | 1.35 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chrX_-_84363974 | 1.35 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr1_-_72566613 | 1.35 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr16_-_57219966 | 1.33 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr6_+_12717892 | 1.32 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_-_81763127 | 1.31 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr4_+_79567233 | 1.31 |
ENST00000514130.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr18_-_70532906 | 1.30 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr12_+_111284764 | 1.27 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr3_+_159943362 | 1.26 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_-_163612840 | 1.24 |
ENST00000419182.1
|
AL078585.1
|
Uncharacterized protein; cDNA FLJ58069 |
| chr1_+_50569575 | 1.23 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_-_87986845 | 1.23 |
ENST00000513893.1
|
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr14_+_62584197 | 1.23 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr1_+_12851545 | 1.23 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr6_+_53883790 | 1.22 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr8_+_7682694 | 1.22 |
ENST00000335186.2
|
DEFB106A
|
defensin, beta 106A |
| chr12_-_11062161 | 1.20 |
ENST00000390677.2
|
TAS2R13
|
taste receptor, type 2, member 13 |
| chr2_-_224467002 | 1.20 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr1_-_13390765 | 1.20 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr20_+_23471727 | 1.19 |
ENST00000449810.1
ENST00000246012.1 |
CST8
|
cystatin 8 (cystatin-related epididymal specific) |
| chr8_-_16859690 | 1.19 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr3_-_51909600 | 1.18 |
ENST00000446461.1
|
IQCF5
|
IQ motif containing F5 |
| chr18_+_61616510 | 1.16 |
ENST00000408945.3
|
HMSD
|
histocompatibility (minor) serpin domain containing |
| chr13_-_45775162 | 1.16 |
ENST00000405872.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr7_-_141541221 | 1.13 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr1_-_13452656 | 1.13 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr17_-_39553844 | 1.11 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr8_-_114449112 | 1.11 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chrX_-_148571884 | 1.10 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr2_+_86333340 | 1.09 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr4_-_76649546 | 1.07 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr12_-_45315625 | 1.05 |
ENST00000552993.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr16_+_32264040 | 1.05 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
| chr10_-_46620012 | 1.02 |
ENST00000508602.1
ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr6_+_97010424 | 1.02 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr16_-_57219926 | 1.02 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr11_-_132812987 | 1.02 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr18_+_5748793 | 1.01 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr7_-_137028498 | 1.01 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chrX_-_138072729 | 1.00 |
ENST00000455663.1
|
FGF13
|
fibroblast growth factor 13 |
| chr15_-_93353028 | 1.00 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr11_-_123525289 | 0.98 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr2_+_96257766 | 0.98 |
ENST00000272395.2
|
TRIM43
|
tripartite motif containing 43 |
| chr7_-_82792215 | 0.97 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr4_-_87028478 | 0.97 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_90049828 | 0.97 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr7_-_137028534 | 0.96 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr15_-_42749711 | 0.95 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr8_+_11225911 | 0.93 |
ENST00000284481.3
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr11_+_125616184 | 0.92 |
ENST00000305738.5
ENST00000437148.2 |
PATE1
|
prostate and testis expressed 1 |
| chr11_-_41481135 | 0.92 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr3_-_128186091 | 0.92 |
ENST00000319153.3
|
DNAJB8
|
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chr4_+_79567057 | 0.91 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chrX_+_64708615 | 0.91 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr5_+_36606700 | 0.91 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_73935163 | 0.90 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr3_+_107364683 | 0.90 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr11_-_89654935 | 0.89 |
ENST00000530311.2
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr5_+_142149955 | 0.89 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr6_-_130182410 | 0.89 |
ENST00000368143.1
|
TMEM244
|
transmembrane protein 244 |
| chr4_+_646960 | 0.88 |
ENST00000488061.1
ENST00000429163.2 |
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr6_-_71666732 | 0.88 |
ENST00000230053.6
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr12_-_119199461 | 0.87 |
ENST00000536572.1
ENST00000542577.1 |
RP11-3L23.2
|
RP11-3L23.2 |
| chr14_+_53019993 | 0.87 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr10_-_48806939 | 0.87 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr2_+_46706725 | 0.86 |
ENST00000434431.1
|
TMEM247
|
transmembrane protein 247 |
| chr7_-_14942944 | 0.86 |
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_90049878 | 0.84 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_+_30856507 | 0.84 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_+_61151306 | 0.83 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_111284805 | 0.83 |
ENST00000552694.1
|
CCDC63
|
coiled-coil domain containing 63 |
| chr6_+_26199737 | 0.82 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr19_+_32836499 | 0.81 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr9_-_107361788 | 0.81 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr9_-_8857776 | 0.81 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr11_-_89540388 | 0.81 |
ENST00000532501.2
|
TRIM49
|
tripartite motif containing 49 |
| chr3_-_39321512 | 0.80 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chrX_+_30260054 | 0.80 |
ENST00000378982.2
|
MAGEB4
|
melanoma antigen family B, 4 |
| chr11_-_123525648 | 0.80 |
ENST00000527836.1
|
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr12_-_112123524 | 0.80 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr2_+_54342574 | 0.79 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr22_+_41956767 | 0.78 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr18_-_74839891 | 0.78 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr10_+_134150835 | 0.78 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr3_-_196242233 | 0.77 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_-_76072719 | 0.77 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_+_126911974 | 0.77 |
ENST00000398112.1
|
C3orf56
|
chromosome 3 open reading frame 56 |
| chr22_+_27043175 | 0.77 |
ENST00000440347.1
ENST00000450203.1 ENST00000430483.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr7_+_123488124 | 0.76 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr12_+_120740119 | 0.76 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr12_-_48963829 | 0.76 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr3_-_164914640 | 0.75 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chrX_-_24690771 | 0.75 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr8_-_18744528 | 0.75 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_-_167098059 | 0.75 |
ENST00000392764.1
ENST00000474464.1 ENST00000392766.2 ENST00000485651.1 |
ZBBX
|
zinc finger, B-box domain containing |
| chr5_+_140227048 | 0.74 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr13_+_109248500 | 0.74 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr7_+_30589829 | 0.74 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr5_-_9712312 | 0.74 |
ENST00000506620.1
ENST00000514078.1 ENST00000606744.1 |
TAS2R1
CTD-2143L24.1
|
taste receptor, type 2, member 1 CTD-2143L24.1 |
| chr2_+_11674213 | 0.74 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_+_168434678 | 0.73 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr5_-_78809950 | 0.73 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr11_+_55650773 | 0.73 |
ENST00000449290.2
|
TRIM51
|
tripartite motif-containing 51 |
| chr16_-_52119019 | 0.72 |
ENST00000561513.1
ENST00000565742.1 |
LINC00919
|
long intergenic non-protein coding RNA 919 |
| chr12_+_127221553 | 0.72 |
ENST00000535118.1
|
LINC00943
|
long intergenic non-protein coding RNA 943 |
| chr2_-_40680578 | 0.71 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_+_12976450 | 0.71 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr8_-_54163972 | 0.71 |
ENST00000520287.1
|
OPRK1
|
opioid receptor, kappa 1 |
| chr4_+_76649797 | 0.70 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr3_-_33759699 | 0.69 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr19_-_14945933 | 0.69 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr16_-_32688053 | 0.69 |
ENST00000398682.4
|
TP53TG3
|
TP53 target 3 |
| chr15_+_62853562 | 0.69 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chrX_+_33744503 | 0.68 |
ENST00000439992.1
|
RP11-305F18.1
|
RP11-305F18.1 |
| chr3_-_47950745 | 0.68 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr3_+_107364769 | 0.68 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr19_-_56709162 | 0.68 |
ENST00000589938.1
ENST00000587032.2 ENST00000586855.2 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr15_+_101402041 | 0.68 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chrX_+_154444643 | 0.67 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr12_+_114514938 | 0.67 |
ENST00000552413.1
|
RP11-100F15.1
|
RP11-100F15.1 |
| chr5_+_98109322 | 0.67 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chrX_+_109602039 | 0.67 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chrX_-_55187588 | 0.67 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr11_-_40315640 | 0.66 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr12_+_72332618 | 0.65 |
ENST00000333850.3
|
TPH2
|
tryptophan hydroxylase 2 |
| chr1_-_13673511 | 0.65 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr8_+_82192501 | 0.65 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr7_-_88425025 | 0.64 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr8_+_7397150 | 0.64 |
ENST00000533250.1
|
RP11-1118M6.1
|
proline rich 23 domain containing 1 |
| chr17_+_61271355 | 0.63 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_68671310 | 0.63 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr5_+_118965244 | 0.63 |
ENST00000515256.1
ENST00000509264.1 |
FAM170A
|
family with sequence similarity 170, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.8 | 5.0 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.7 | 2.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.4 | 1.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 2.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 1.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.3 | 3.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 6.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 0.8 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.3 | 1.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 0.8 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.3 | 0.8 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.3 | 1.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.4 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.7 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.2 | 1.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.2 | 1.8 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.2 | 1.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 2.7 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 | 2.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.8 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.7 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 3.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.8 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.8 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.5 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 1.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.6 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.6 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 4.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 0.7 | GO:0042435 | serotonin biosynthetic process(GO:0042427) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.1 | 2.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 3.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 7.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 2.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 3.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.1 | 2.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 1.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 1.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 3.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:0070293 | renal sodium ion transport(GO:0003096) renal absorption(GO:0070293) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.5 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 2.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 1.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 2.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 2.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.5 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 2.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.5 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 2.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 1.9 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.8 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 1.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.0 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.6 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 2.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0007154 | cell communication(GO:0007154) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.0 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 3.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.9 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.2 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 4.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 2.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 3.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.4 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 1.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 3.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 4.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 5.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 3.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 10.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 3.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 4.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 3.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.1 | GO:1990904 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.5 | 2.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.4 | 11.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 1.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 1.1 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.3 | 1.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 3.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 3.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 0.8 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.2 | 2.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.7 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.2 | 1.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 1.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 2.7 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 0.8 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 1.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 4.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 5.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 2.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 2.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.5 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 1.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 3.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 2.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 4.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 12.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 3.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 6.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 4.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |