Project
Illumina Body Map 2
Navigation
Downloads
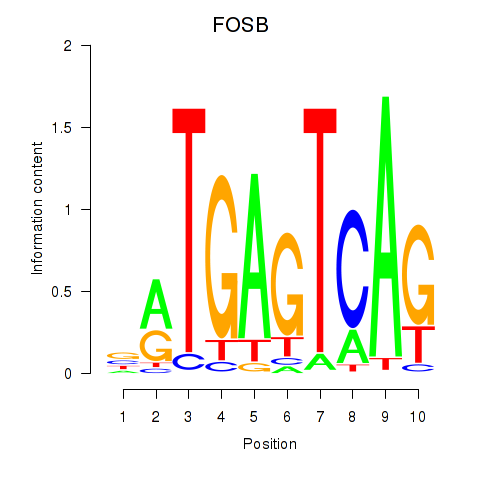
Results for FOSB
Z-value: 0.94
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.9 | FosB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSB | hg19_v2_chr19_+_45971246_45971265 | -0.30 | 9.8e-02 | Click! |
Activity profile of FOSB motif
Sorted Z-values of FOSB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_35685113 | 1.97 |
ENST00000419330.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_10920714 | 1.88 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr17_+_4853442 | 1.88 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_-_220173685 | 1.84 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr2_+_27505260 | 1.65 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr17_-_8021710 | 1.57 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr1_-_190443931 | 1.57 |
ENST00000445957.2
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr14_-_75083313 | 1.55 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr10_-_76818272 | 1.54 |
ENST00000338487.5
|
DUPD1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr11_-_64527425 | 1.52 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr5_-_176923803 | 1.51 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr6_-_24877490 | 1.49 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr2_-_97509749 | 1.46 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr11_-_10920838 | 1.43 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr11_+_10477733 | 1.43 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_-_97509729 | 1.40 |
ENST00000418232.1
|
ANKRD23
|
ankyrin repeat domain 23 |
| chr6_-_113953705 | 1.36 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr10_-_75423560 | 1.36 |
ENST00000606523.1
|
SYNPO2L
|
synaptopodin 2-like |
| chr17_-_33390667 | 1.32 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_-_176923846 | 1.32 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr1_-_21059029 | 1.28 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr18_+_74207477 | 1.19 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr21_+_30672433 | 1.16 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr1_+_223889285 | 1.13 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr5_+_175288631 | 1.13 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr19_-_35981358 | 1.12 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr11_-_102714534 | 1.11 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr17_-_59668550 | 1.09 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr17_-_27503770 | 1.09 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_66103867 | 1.09 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_-_3595042 | 1.07 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr16_-_72206034 | 1.05 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr3_+_184534994 | 1.03 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr10_+_48247669 | 1.01 |
ENST00000457620.1
|
FAM25G
|
family with sequence similarity 25, member G |
| chr11_-_71781096 | 1.01 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr10_-_47181681 | 1.00 |
ENST00000452267.1
|
FAM25B
|
family with sequence similarity 25, member B |
| chr1_+_22333943 | 1.00 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr22_+_21369316 | 0.99 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr11_-_66103932 | 0.98 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr12_-_95010147 | 0.98 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr6_-_41673552 | 0.98 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr6_+_292051 | 0.97 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr18_-_74207146 | 0.96 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr3_+_183894566 | 0.95 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr17_+_79650962 | 0.95 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr17_-_73150629 | 0.95 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr4_+_84457529 | 0.95 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr16_+_30675654 | 0.95 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr18_+_9885760 | 0.95 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr4_+_71588372 | 0.94 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_-_80329356 | 0.93 |
ENST00000358842.3
|
GK2
|
glycerol kinase 2 |
| chr16_+_56782118 | 0.92 |
ENST00000566678.1
|
NUP93
|
nucleoporin 93kDa |
| chr17_+_80317121 | 0.91 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr17_-_27467418 | 0.91 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr18_+_9885961 | 0.90 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr12_-_95009837 | 0.88 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr4_+_41983713 | 0.88 |
ENST00000333141.5
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1 |
| chr3_-_196065374 | 0.87 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr11_-_65150103 | 0.87 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr4_+_84457250 | 0.86 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr16_+_56781814 | 0.86 |
ENST00000568656.1
|
NUP93
|
nucleoporin 93kDa |
| chr20_-_634000 | 0.85 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr20_+_21686290 | 0.84 |
ENST00000398485.2
|
PAX1
|
paired box 1 |
| chr4_+_170581213 | 0.84 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr11_-_46848393 | 0.84 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr12_-_96389702 | 0.83 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr6_-_35888905 | 0.81 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_+_155537771 | 0.80 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_155538093 | 0.79 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr11_+_63448918 | 0.79 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr11_-_9781068 | 0.78 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr17_+_25799008 | 0.77 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr6_-_127837739 | 0.77 |
ENST00000368268.2
|
SOGA3
|
SOGA family member 3 |
| chr19_-_18995029 | 0.75 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr8_-_42623747 | 0.74 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_-_160231451 | 0.74 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_-_235116495 | 0.73 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr3_+_183894737 | 0.73 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr17_-_18908040 | 0.72 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr22_-_39151434 | 0.72 |
ENST00000439339.1
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr10_+_88780049 | 0.71 |
ENST00000343959.4
|
FAM25A
|
family with sequence similarity 25, member A |
| chr9_-_35112376 | 0.71 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr18_+_9885934 | 0.70 |
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr11_-_123756334 | 0.70 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr17_+_21191341 | 0.70 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr10_+_134901388 | 0.69 |
ENST00000392607.3
|
GPR123
|
G protein-coupled receptor 123 |
| chr13_-_41768654 | 0.69 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr17_-_18161870 | 0.68 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr15_+_67458357 | 0.67 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr19_-_36019123 | 0.67 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_-_179112189 | 0.67 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_+_208104497 | 0.66 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr7_+_142919130 | 0.66 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr15_+_67418047 | 0.66 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr10_-_111713633 | 0.66 |
ENST00000538668.1
ENST00000369657.1 ENST00000369655.1 |
RP11-451M19.3
|
Uncharacterized protein |
| chr14_-_96180435 | 0.66 |
ENST00000556450.1
ENST00000555202.1 ENST00000554012.1 ENST00000402399.1 |
TCL1A
|
T-cell leukemia/lymphoma 1A |
| chr5_+_145317356 | 0.65 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr6_+_108487245 | 0.65 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr5_+_143191726 | 0.64 |
ENST00000289448.2
|
HMHB1
|
histocompatibility (minor) HB-1 |
| chr1_+_101003687 | 0.64 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr19_+_38880695 | 0.64 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chrX_-_154493791 | 0.64 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr9_-_114246635 | 0.63 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr12_-_96390063 | 0.63 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr16_+_89988259 | 0.63 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr3_-_18466787 | 0.63 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr9_+_137218362 | 0.62 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr6_+_159290917 | 0.62 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr18_-_40695604 | 0.62 |
ENST00000590910.1
ENST00000326695.5 ENST00000589109.1 ENST00000282028.4 |
RIT2
|
Ras-like without CAAX 2 |
| chr12_-_45315625 | 0.62 |
ENST00000552993.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr8_-_125384927 | 0.61 |
ENST00000297632.6
|
TMEM65
|
transmembrane protein 65 |
| chr17_+_30771279 | 0.60 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr8_+_123875624 | 0.60 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr2_-_119605253 | 0.59 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr8_-_17752996 | 0.58 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr11_-_94965667 | 0.58 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr16_-_4664860 | 0.58 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr9_+_97562440 | 0.57 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr12_+_6644443 | 0.57 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_+_35851570 | 0.57 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr17_-_72864739 | 0.56 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr8_-_110988070 | 0.55 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr17_+_75315534 | 0.55 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr9_+_34992846 | 0.55 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr16_-_80603558 | 0.55 |
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1 |
| chr5_-_61031495 | 0.54 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr14_-_74226961 | 0.54 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr12_+_53693466 | 0.54 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr19_+_926000 | 0.53 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr17_+_75446819 | 0.53 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr17_-_28257080 | 0.52 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr12_+_52695617 | 0.52 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_-_93257951 | 0.51 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr3_+_48507210 | 0.51 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr6_-_35888824 | 0.51 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr12_-_96390108 | 0.51 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr1_-_6662919 | 0.51 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr7_-_142120321 | 0.50 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr3_+_113465866 | 0.50 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr10_+_121410882 | 0.50 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr13_-_52027134 | 0.50 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr3_+_48507621 | 0.50 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr3_-_18466026 | 0.50 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr2_-_219858123 | 0.49 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chrX_+_133371077 | 0.49 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr12_+_58176525 | 0.49 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr3_+_108308845 | 0.49 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr17_-_27405875 | 0.49 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr15_+_75335604 | 0.48 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr4_+_120056939 | 0.48 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr2_-_145275109 | 0.48 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_33765593 | 0.48 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr22_-_32860427 | 0.48 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chr10_-_29923893 | 0.47 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr17_-_38911580 | 0.47 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr12_+_28605426 | 0.46 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr15_+_45938079 | 0.46 |
ENST00000561493.1
|
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr5_-_41794663 | 0.46 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr16_-_4665023 | 0.46 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr1_-_207224307 | 0.46 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr11_-_65430251 | 0.46 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr2_+_27237615 | 0.45 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr1_-_236046872 | 0.45 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr17_-_7123021 | 0.45 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr3_-_51994694 | 0.44 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr2_-_96926313 | 0.44 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr21_+_19617140 | 0.44 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr11_-_124180733 | 0.44 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1 |
| chr17_+_76311791 | 0.43 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr12_+_7023491 | 0.43 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr19_-_46145696 | 0.43 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr3_-_131756559 | 0.42 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr8_-_42623924 | 0.42 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr1_+_154975258 | 0.42 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr9_+_131902346 | 0.42 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr8_-_17752912 | 0.42 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr14_+_31494672 | 0.41 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_-_75201791 | 0.40 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr12_-_118628350 | 0.40 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr10_+_68685764 | 0.40 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr7_-_102985035 | 0.40 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr19_-_55881741 | 0.39 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr16_-_67281413 | 0.39 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr5_+_159436120 | 0.38 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr16_-_1429627 | 0.38 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr20_+_48429356 | 0.38 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr3_+_157828152 | 0.38 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr6_+_2988847 | 0.37 |
ENST00000380472.3
ENST00000605901.1 ENST00000454015.1 |
NQO2
LINC01011
|
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr11_+_68671310 | 0.37 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr14_+_97925151 | 0.37 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr5_-_41794313 | 0.37 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr6_-_35888858 | 0.37 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr1_+_17634689 | 0.36 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr11_-_47206965 | 0.36 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr10_-_86001210 | 0.36 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr1_+_110026544 | 0.36 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr12_+_55968199 | 0.36 |
ENST00000321688.1
|
OR2AP1
|
olfactory receptor, family 2, subfamily AP, member 1 |
| chr20_+_48429233 | 0.36 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr1_-_153029980 | 0.36 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr16_-_1429674 | 0.36 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.5 | 1.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 0.9 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 2.0 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.7 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.2 | 1.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 2.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.8 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 1.6 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.5 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.2 | 0.5 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 2.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 2.9 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 0.4 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.9 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.6 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 1.0 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 2.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.3 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.1 | 0.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 1.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 3.0 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 1.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 2.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 1.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.7 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:2000321 | interleukin-18 production(GO:0032621) positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.7 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 1.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 1.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0032899 | regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 1.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0048631 | negative regulation of skeletal muscle cell proliferation(GO:0014859) skeletal muscle tissue growth(GO:0048630) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 1.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.5 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.3 | 2.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 1.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.2 | 2.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 1.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.5 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 3.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.0 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 5.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 3.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 2.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.7 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.4 | 1.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.6 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.3 | 0.9 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 2.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 1.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.6 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.2 | 2.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 2.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 1.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.3 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.4 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.2 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 3.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |