Project
Illumina Body Map 2
Navigation
Downloads
Results for FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 1.46
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
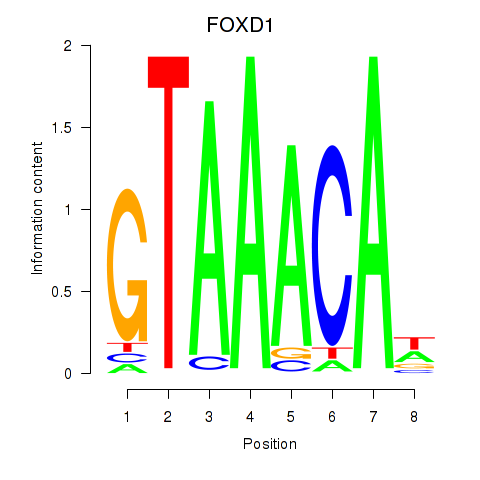
FOXD1
|
ENSG00000251493.2 | forkhead box D1 |
|
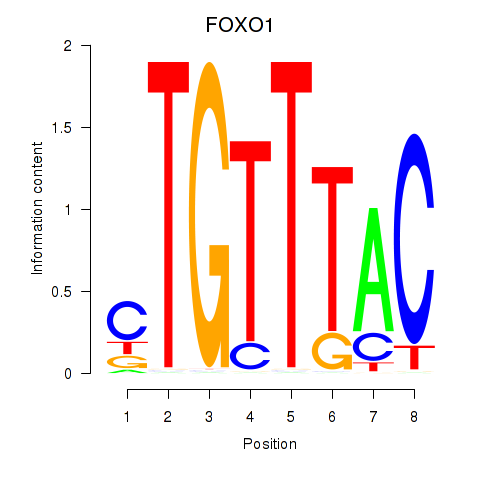
FOXO1
|
ENSG00000150907.6 | forkhead box O1 |
|
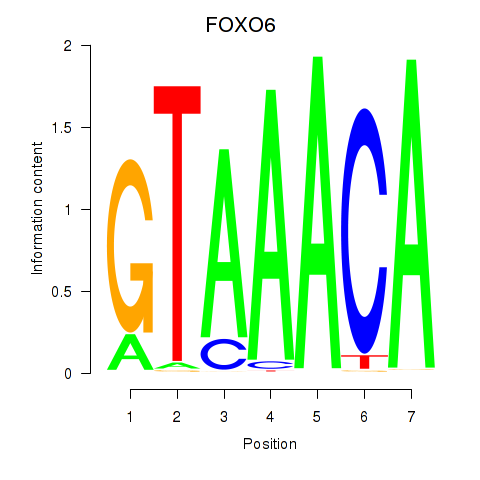
FOXO6
|
ENSG00000204060.4 | forkhead box O6 |
|
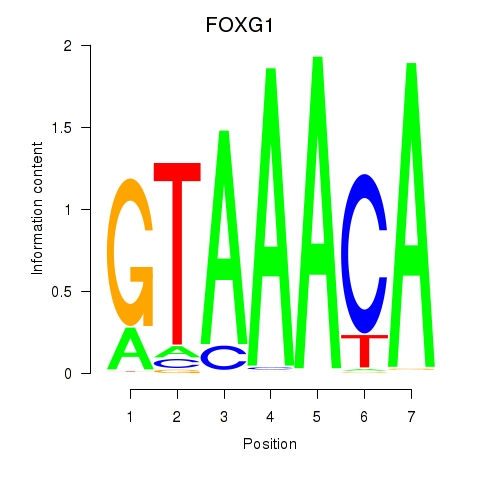
FOXG1
|
ENSG00000176165.7 | forkhead box G1 |
|
FOXP1
|
ENSG00000114861.14 | forkhead box P1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXG1 | hg19_v2_chr14_+_29234870_29235050 | -0.61 | 2.3e-04 | Click! |
| FOXO1 | hg19_v2_chr13_-_41240717_41240735 | 0.42 | 1.6e-02 | Click! |
| FOXP1 | hg19_v2_chr3_-_71179699_71179744 | 0.15 | 4.1e-01 | Click! |
| FOXO6 | hg19_v2_chr1_+_41827594_41827594 | -0.12 | 5.1e-01 | Click! |
| FOXD1 | hg19_v2_chr5_-_72744336_72744359 | -0.04 | 8.4e-01 | Click! |
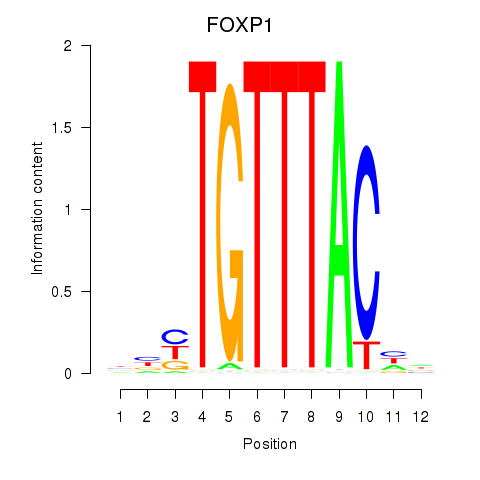
Activity profile of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
Sorted Z-values of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_145438469 | 5.60 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr10_+_63808970 | 5.44 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr18_+_3449330 | 5.36 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr21_+_39668478 | 4.77 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_-_95225768 | 4.74 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr7_+_134832808 | 4.38 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chrX_+_135251835 | 3.81 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135251783 | 3.78 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr21_+_39668831 | 3.74 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_-_168865522 | 3.52 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_-_26394114 | 3.50 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr17_-_1418972 | 3.48 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr13_-_46716969 | 3.42 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_-_1508379 | 3.37 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chrX_+_117629766 | 3.21 |
ENST00000276204.6
ENST00000276202.7 |
DOCK11
|
dedicator of cytokinesis 11 |
| chr6_-_42016385 | 3.12 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr10_+_115469134 | 3.09 |
ENST00000452490.2
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr14_-_36988882 | 2.98 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr20_-_50179368 | 2.96 |
ENST00000609943.1
ENST00000609507.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr8_+_126442563 | 2.88 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chrX_+_135252050 | 2.88 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr19_-_18902106 | 2.88 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr17_-_39942940 | 2.86 |
ENST00000310706.5
ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP
|
junction plakoglobin |
| chr17_-_10450866 | 2.84 |
ENST00000578017.1
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr12_-_92539614 | 2.77 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr11_+_117049910 | 2.77 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr3_+_148447887 | 2.71 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr10_+_123923105 | 2.66 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_-_116714564 | 2.66 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chrX_-_106960285 | 2.64 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr18_+_3449695 | 2.58 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_+_117049854 | 2.56 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_+_10476851 | 2.54 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr19_+_12902289 | 2.51 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr1_+_207669573 | 2.47 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr12_+_13349650 | 2.42 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr22_-_31688381 | 2.40 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr12_+_54378923 | 2.39 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr9_+_134165195 | 2.37 |
ENST00000372261.1
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr17_-_42345487 | 2.34 |
ENST00000262418.6
|
SLC4A1
|
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr5_+_72251857 | 2.33 |
ENST00000507345.2
ENST00000512348.1 ENST00000287761.6 |
FCHO2
|
FCH domain only 2 |
| chr1_+_207669613 | 2.32 |
ENST00000367049.4
ENST00000529814.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr17_-_46657473 | 2.30 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr12_+_13349711 | 2.30 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr7_+_80231466 | 2.30 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_227127981 | 2.25 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr10_+_104535994 | 2.24 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr14_-_23288930 | 2.23 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr22_-_31688431 | 2.23 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_152297679 | 2.22 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr12_+_54378849 | 2.21 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr11_+_844406 | 2.21 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr8_+_128747661 | 2.18 |
ENST00000259523.6
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr6_-_31550192 | 2.17 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr12_-_76817036 | 2.16 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_-_36983034 | 2.16 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr17_-_8059638 | 2.15 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr19_-_42916499 | 2.14 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr14_-_54418598 | 2.14 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr6_+_108977520 | 2.09 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr11_-_111781554 | 2.09 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chrX_-_15619076 | 2.07 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_+_89791507 | 2.06 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr13_-_41240717 | 2.03 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr2_+_111878483 | 2.02 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr7_+_30174574 | 2.01 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_+_77469439 | 2.01 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr14_+_23067166 | 2.00 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr19_+_18496957 | 1.99 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr11_+_844067 | 1.99 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr4_-_141075330 | 1.99 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr18_-_53257027 | 1.95 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr9_+_134165063 | 1.94 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr19_-_39826639 | 1.93 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr11_-_111781610 | 1.92 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_150460539 | 1.91 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_111781454 | 1.90 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_142782862 | 1.89 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr10_+_75668916 | 1.89 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr16_+_57680840 | 1.89 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr3_-_71294304 | 1.88 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr7_+_106809406 | 1.87 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr18_+_9475585 | 1.87 |
ENST00000585015.1
|
RALBP1
|
ralA binding protein 1 |
| chr3_+_69985792 | 1.86 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_-_46608010 | 1.86 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr1_+_152486950 | 1.86 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr3_+_69985734 | 1.85 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr8_+_97773202 | 1.85 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr8_+_128747757 | 1.84 |
ENST00000517291.1
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr13_+_28712614 | 1.84 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_-_45909585 | 1.83 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_+_57680811 | 1.82 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr8_+_128748466 | 1.81 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr1_-_12679171 | 1.80 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr2_-_214016314 | 1.80 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_-_28503693 | 1.80 |
ENST00000373857.3
|
PTAFR
|
platelet-activating factor receptor |
| chr11_+_117049445 | 1.78 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr8_+_128748308 | 1.76 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr8_+_99956662 | 1.76 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr7_-_84122033 | 1.76 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_12677714 | 1.75 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr19_+_13906250 | 1.75 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr8_-_133772794 | 1.74 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr8_-_124553437 | 1.74 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr14_+_56127960 | 1.73 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_-_16509101 | 1.72 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr12_-_29936731 | 1.72 |
ENST00000552618.1
ENST00000539277.1 ENST00000551659.1 |
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr15_-_60884706 | 1.71 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr8_-_29208183 | 1.71 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr7_-_27213893 | 1.71 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr14_+_24584372 | 1.70 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr19_-_47734448 | 1.67 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr12_+_106696581 | 1.66 |
ENST00000547153.1
ENST00000299045.3 ENST00000546625.1 ENST00000553098.1 |
TCP11L2
|
t-complex 11, testis-specific-like 2 |
| chr1_-_207095324 | 1.63 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr6_+_74405501 | 1.63 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr12_+_54410664 | 1.62 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr5_+_40841276 | 1.62 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr16_+_57679859 | 1.60 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_-_140624499 | 1.59 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr3_-_71179988 | 1.58 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr12_-_53045948 | 1.58 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr2_-_160472952 | 1.57 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_152149033 | 1.56 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr10_-_14050522 | 1.56 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr7_-_115608304 | 1.54 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr5_+_40841410 | 1.53 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_+_171154347 | 1.53 |
ENST00000209929.7
ENST00000441535.1 |
FMO2
|
flavin containing monooxygenase 2 (non-functional) |
| chr1_-_207095212 | 1.52 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr20_-_49308048 | 1.52 |
ENST00000327979.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr4_-_174256276 | 1.52 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr17_+_26662679 | 1.51 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr7_+_28725585 | 1.51 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr8_-_133772870 | 1.50 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr8_+_97773457 | 1.50 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr17_-_6947225 | 1.49 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr5_-_16742330 | 1.49 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr16_+_4896659 | 1.49 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr6_+_74405804 | 1.48 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr9_-_115095123 | 1.48 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr15_-_70994612 | 1.48 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr14_+_56127989 | 1.47 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr9_-_20382446 | 1.47 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr16_+_53242350 | 1.47 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_169725231 | 1.46 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr7_-_27219849 | 1.46 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr16_+_57679945 | 1.45 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr18_-_53070913 | 1.44 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr5_+_72251793 | 1.44 |
ENST00000430046.2
ENST00000341845.6 |
FCHO2
|
FCH domain only 2 |
| chr7_-_37026108 | 1.43 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_+_109223595 | 1.43 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_-_7293942 | 1.43 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr14_+_24584508 | 1.42 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_17980091 | 1.42 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr18_-_59415987 | 1.41 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr9_+_27109133 | 1.41 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr1_+_116654376 | 1.40 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr11_-_57089774 | 1.40 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr16_-_57831914 | 1.38 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr7_+_30174426 | 1.38 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr9_+_112852477 | 1.38 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr16_+_57680043 | 1.37 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr5_+_102200948 | 1.37 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_24584056 | 1.37 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_134832752 | 1.35 |
ENST00000452718.1
|
AC083862.1
|
Uncharacterized protein |
| chr8_+_92261516 | 1.34 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr11_+_3968573 | 1.34 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr9_+_135457530 | 1.34 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr4_-_40632605 | 1.34 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_+_79369249 | 1.33 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr15_-_34629922 | 1.33 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr2_+_97481974 | 1.32 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr3_-_71114066 | 1.32 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr4_-_74486217 | 1.32 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_123187890 | 1.32 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr13_-_31038370 | 1.32 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr20_-_49307897 | 1.31 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr17_-_46035187 | 1.31 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr14_-_36989336 | 1.31 |
ENST00000522719.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_-_57832004 | 1.30 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr17_+_33474826 | 1.30 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr3_+_188889737 | 1.29 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr17_+_26662597 | 1.29 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr11_-_57089671 | 1.29 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr1_+_66796401 | 1.29 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_-_84150392 | 1.28 |
ENST00000570012.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr17_-_40897043 | 1.27 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
| chr17_+_33474860 | 1.27 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr6_-_152623231 | 1.27 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr7_-_5463175 | 1.27 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr1_+_198126093 | 1.27 |
ENST00000367385.4
ENST00000442588.1 ENST00000538004.1 |
NEK7
|
NIMA-related kinase 7 |
| chr16_-_57831676 | 1.26 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr9_-_128246769 | 1.25 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr12_-_123201337 | 1.25 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr12_-_53074182 | 1.25 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr9_+_27109392 | 1.25 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr12_+_96588279 | 1.25 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr8_+_52730143 | 1.24 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr4_-_40632881 | 1.24 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr19_-_50380536 | 1.23 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr9_-_115095229 | 1.23 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 1.6 | 4.8 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 1.2 | 7.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 1.0 | 3.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.0 | 2.9 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.9 | 4.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.8 | 2.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.7 | 3.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 2.7 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.6 | 0.6 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.6 | 1.8 | GO:1904301 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.6 | 2.9 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.6 | 3.5 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.6 | 2.8 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 1.9 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.5 | 3.4 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.5 | 3.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 7.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 1.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 2.2 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.4 | 2.1 | GO:0072097 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 | 3.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 2.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.4 | 2.9 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.4 | 2.0 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.4 | 3.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 1.6 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.4 | 2.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 1.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.4 | 2.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.4 | 2.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.4 | 1.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.4 | 4.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 3.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 1.0 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.3 | 1.3 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.3 | 1.0 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.3 | 1.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.3 | 2.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 2.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 3.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 2.4 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.3 | 0.3 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.3 | 2.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.3 | 4.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 1.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.3 | 2.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 2.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.3 | 0.5 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.3 | 1.1 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.3 | 2.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 3.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 2.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.0 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.2 | 1.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 1.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.0 | GO:0003274 | endocardial cushion fusion(GO:0003274) bronchus morphogenesis(GO:0060434) |
| 0.2 | 1.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 2.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 3.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 1.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 6.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 4.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 1.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.9 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.2 | 0.7 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 2.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 5.6 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 2.2 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 2.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 3.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 3.8 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 5.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 3.4 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 0.6 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 2.3 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 4.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 3.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 2.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) |
| 0.2 | 0.6 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.2 | 12.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 2.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 0.7 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.2 | 2.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 | 1.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 2.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 10.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 0.5 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.2 | 2.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 2.8 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 0.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.5 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.2 | 2.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 1.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.6 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.8 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.4 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.9 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 2.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 1.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.9 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 1.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.3 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 3.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 2.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.4 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.5 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.9 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 1.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.8 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.8 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 1.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.8 | GO:0032455 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) nerve growth factor processing(GO:0032455) |
| 0.1 | 0.3 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.1 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.9 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.6 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.7 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.3 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.7 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.5 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 3.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 3.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 3.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 3.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 4.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.1 | 0.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 4.7 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.4 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.3 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 5.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.7 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 1.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.3 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 1.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.7 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.0 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.5 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.5 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 4.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 1.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 2.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.5 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.1 | 0.4 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 3.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.1 | 0.2 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.5 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.8 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.3 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 0.2 | GO:1903980 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.3 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 2.3 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.0 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 2.6 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 0.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.5 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 1.1 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.3 | GO:1901355 | cellular response to rapamycin(GO:0072752) response to rapamycin(GO:1901355) |
| 0.0 | 1.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 1.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 1.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.2 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 2.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.9 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 1.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 1.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.5 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 3.5 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 0.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.6 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 1.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 1.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.5 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.7 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.6 | GO:0017038 | protein import into nucleus(GO:0006606) protein import(GO:0017038) protein targeting to nucleus(GO:0044744) |
| 0.0 | 1.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.5 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 2.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 1.0 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 1.2 | GO:1900740 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 2.7 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 1.4 | GO:0043367 | CD4-positive, alpha-beta T cell differentiation(GO:0043367) |
| 0.0 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.4 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 4.4 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.5 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0042321 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) small intestine smooth muscle contraction(GO:1990770) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.1 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 0.1 | GO:2001214 | regulation of vasculogenesis(GO:2001212) positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 1.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 6.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.5 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.7 | 2.9 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.6 | 2.9 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.5 | 2.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.5 | 1.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 0.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 2.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 4.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 2.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 5.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 2.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 1.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 3.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 2.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 1.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 1.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 2.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 6.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 5.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 1.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 3.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 2.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.3 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 11.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.1 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 3.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 6.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.6 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 3.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 4.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 8.8 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 2.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 6.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 26.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 2.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 6.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 4.7 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.4 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 2.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 8.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 7.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 56.1 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 1.5 | 4.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.2 | 4.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.2 | 7.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 2.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.7 | 2.9 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.7 | 2.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.7 | 2.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.7 | 2.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 1.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 10.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 3.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.4 | 1.3 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.4 | 3.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.4 | 2.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 5.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 3.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.3 | 1.5 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.3 | 23.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 1.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.3 | 0.8 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.3 | 0.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 1.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.3 | 2.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.8 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.3 | 3.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.2 | 2.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 0.9 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 8.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 2.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 5.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 0.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 0.8 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 4.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.6 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.2 | 1.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 4.0 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.7 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 3.6 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.8 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 1.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.5 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 3.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 3.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 1.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.9 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 2.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 8.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 3.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 4.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.9 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 9.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 3.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 3.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 3.3 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 2.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 5.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 3.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 2.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.8 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.1 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 3.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 1.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 4.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 2.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 1.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.3 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.4 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 2.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 4.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 4.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 3.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 4.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 2.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.0 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 2.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 3.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.9 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 2.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.0 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 3.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 11.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 10.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 5.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 15.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 16.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 8.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 5.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 4.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 11.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 4.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 5.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 10.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.2 | 1.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 8.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 3.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 8.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.6 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 1.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 6.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 5.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 3.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.7 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.1 | 4.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 3.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 6.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 4.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 2.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 9.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |