Project
Illumina Body Map 2
Navigation
Downloads
Results for FOXO4
Z-value: 1.42
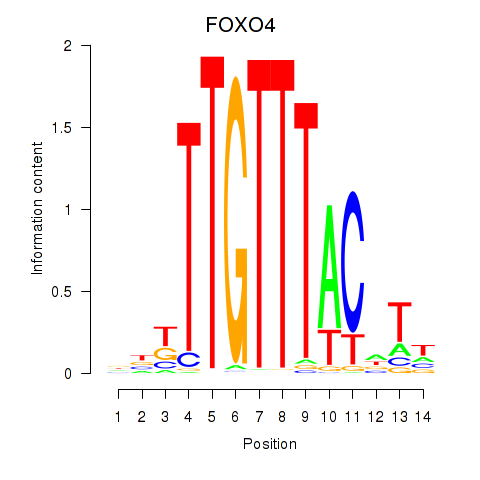
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | 0.20 | 2.8e-01 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_105627735 | 4.88 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr12_-_111358372 | 4.34 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr7_-_95225768 | 4.25 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr10_+_123923105 | 4.08 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr10_-_4285835 | 3.98 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr10_-_4285923 | 3.76 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr11_+_113930955 | 3.40 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr10_-_13570533 | 3.32 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr10_+_123923205 | 3.31 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_100242549 | 3.20 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr4_-_186696425 | 3.19 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_32964258 | 3.18 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_86934526 | 3.18 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr10_+_123922941 | 3.15 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_97010424 | 3.13 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr4_-_152149033 | 3.09 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr3_+_148447887 | 3.05 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr5_+_137203465 | 2.99 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr5_+_137203557 | 2.87 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr5_+_137203541 | 2.86 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr16_+_6069072 | 2.77 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_+_6069586 | 2.76 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_+_49984894 | 2.76 |
ENST00000522267.1
ENST00000399653.4 ENST00000303202.8 |
C8orf22
|
chromosome 8 open reading frame 22 |
| chr9_+_27109440 | 2.74 |
ENST00000519080.1
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr12_+_53491220 | 2.66 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr17_+_72426891 | 2.57 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr17_+_33474826 | 2.51 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr17_+_33474860 | 2.46 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr17_+_8924837 | 2.46 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr9_+_27109392 | 2.45 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr7_+_134551583 | 2.44 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr2_+_173955327 | 2.33 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr3_-_52868931 | 2.32 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr12_+_54378923 | 2.31 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr6_+_62284008 | 2.27 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr2_-_71454185 | 2.26 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr12_+_54378849 | 2.23 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr4_+_124320665 | 2.23 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chrX_-_38080077 | 2.18 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr8_-_6420565 | 2.18 |
ENST00000338312.6
|
ANGPT2
|
angiopoietin 2 |
| chr12_-_71031185 | 2.15 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_187461442 | 2.11 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr4_-_186696561 | 2.11 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_119247004 | 2.07 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr5_-_59481406 | 2.07 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_7580103 | 2.04 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr6_-_46889694 | 2.03 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr4_-_186696515 | 2.02 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_183731882 | 2.00 |
ENST00000295113.4
|
FRZB
|
frizzled-related protein |
| chr12_-_102872317 | 1.99 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr3_+_159570722 | 1.95 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr17_+_72427477 | 1.94 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_52869205 | 1.92 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr8_-_6420777 | 1.90 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr9_+_27109133 | 1.89 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr7_-_134832752 | 1.88 |
ENST00000452718.1
|
AC083862.1
|
Uncharacterized protein |
| chr17_-_67138015 | 1.84 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr7_+_134832808 | 1.81 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr17_-_67057114 | 1.79 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr18_+_32073253 | 1.79 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr19_+_13134772 | 1.75 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_8075693 | 1.71 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr8_-_6420930 | 1.71 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr18_-_21852143 | 1.69 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr10_-_118032697 | 1.67 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr14_-_61124977 | 1.65 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr7_+_55177416 | 1.64 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr10_+_86184676 | 1.63 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr9_-_95186739 | 1.63 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr17_-_67057047 | 1.63 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_-_57045228 | 1.61 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr3_-_185826855 | 1.59 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr20_+_44035200 | 1.57 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_+_94577074 | 1.56 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr4_-_186697044 | 1.55 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_132316081 | 1.53 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr8_-_13134045 | 1.53 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr5_-_10308125 | 1.50 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr1_-_12679171 | 1.49 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr19_+_859654 | 1.48 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr1_+_200011711 | 1.47 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_-_58522880 | 1.47 |
ENST00000474098.1
|
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr1_-_115238207 | 1.46 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr10_-_37891859 | 1.46 |
ENST00000544824.1
|
MTRNR2L7
|
MT-RNR2-like 7 |
| chr13_-_51101468 | 1.45 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr2_+_233527443 | 1.43 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr18_-_68004529 | 1.40 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr6_-_127780510 | 1.40 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr12_-_71031220 | 1.39 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr19_+_859425 | 1.37 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr9_+_5510492 | 1.37 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr13_-_40924439 | 1.35 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr5_-_59064458 | 1.35 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_+_37723420 | 1.32 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr18_+_28898052 | 1.31 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr1_-_182360498 | 1.30 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr9_+_5510558 | 1.27 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr3_+_138068051 | 1.27 |
ENST00000474559.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr9_+_706842 | 1.25 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr3_+_28390637 | 1.25 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr11_-_114466471 | 1.25 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr11_-_89224299 | 1.25 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr17_-_17875688 | 1.24 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr9_-_140444814 | 1.23 |
ENST00000277531.4
|
PNPLA7
|
patatin-like phospholipase domain containing 7 |
| chr3_-_71777824 | 1.22 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr10_-_118032979 | 1.22 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr9_-_140444867 | 1.22 |
ENST00000406427.1
|
PNPLA7
|
patatin-like phospholipase domain containing 7 |
| chr8_-_6420759 | 1.20 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr11_-_89224139 | 1.16 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr2_+_170683979 | 1.15 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr9_-_13175823 | 1.14 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr19_-_7293942 | 1.14 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr11_-_89223883 | 1.14 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr5_-_42811986 | 1.14 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr10_-_61900762 | 1.13 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_-_182360918 | 1.13 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr22_+_44427230 | 1.12 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr11_-_114466477 | 1.09 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr18_+_6834472 | 1.08 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr1_+_145525015 | 1.07 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr2_+_170683942 | 1.06 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr6_+_108882069 | 1.06 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr2_+_135809835 | 1.06 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr1_-_234667504 | 1.06 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr1_+_63989004 | 1.06 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr10_-_90751038 | 1.05 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr12_-_10251539 | 1.04 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr2_+_182850551 | 1.04 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr17_-_1418972 | 1.03 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr11_+_77532155 | 1.03 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr8_-_123793048 | 1.02 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr9_-_86432547 | 1.02 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr5_-_42812143 | 1.02 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_-_27702455 | 1.02 |
ENST00000265395.2
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase |
| chr10_+_75668916 | 1.01 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr14_+_56127989 | 1.01 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr2_-_100925967 | 1.01 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr11_+_112832090 | 1.01 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr5_-_58571935 | 1.00 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_-_31661000 | 0.99 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr2_+_58134756 | 0.99 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr19_-_50380536 | 0.98 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr1_+_47533160 | 0.97 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr19_+_18111927 | 0.97 |
ENST00000379656.3
|
ARRDC2
|
arrestin domain containing 2 |
| chr12_-_7596735 | 0.95 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr8_-_62602327 | 0.94 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr2_-_23747214 | 0.94 |
ENST00000430988.1
|
AC011239.1
|
Uncharacterized protein |
| chr15_+_63188009 | 0.93 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr19_+_50380917 | 0.93 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr17_-_8263538 | 0.91 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr19_-_42916499 | 0.91 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr20_-_62582475 | 0.89 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr3_-_145940126 | 0.89 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr11_+_77532233 | 0.87 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr3_-_134092561 | 0.87 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr15_-_52971544 | 0.85 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr12_-_10251603 | 0.85 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr6_+_72596604 | 0.84 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_86422523 | 0.83 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr3_-_185826718 | 0.83 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr16_+_84801852 | 0.83 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr7_+_120628731 | 0.82 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_14980632 | 0.82 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr3_+_121289551 | 0.81 |
ENST00000334384.3
|
ARGFX
|
arginine-fifty homeobox |
| chr3_+_112930387 | 0.80 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_+_65283529 | 0.79 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr11_+_34999328 | 0.79 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr15_+_86805875 | 0.79 |
ENST00000389298.3
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr6_+_123110465 | 0.79 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr5_+_98104978 | 0.77 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chr4_-_186696636 | 0.77 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_28857677 | 0.77 |
ENST00000313511.3
|
TUFM
|
Tu translation elongation factor, mitochondrial |
| chr3_+_173302222 | 0.77 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr4_-_140223614 | 0.75 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr14_+_24439148 | 0.75 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr2_-_225434538 | 0.75 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr2_-_166060382 | 0.74 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr15_-_31283618 | 0.74 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr1_+_214163033 | 0.73 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr10_+_111985837 | 0.73 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr10_-_62332357 | 0.72 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_+_67410832 | 0.72 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr6_+_168418553 | 0.72 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chr3_+_124223586 | 0.72 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr2_+_157292859 | 0.71 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr19_-_29704448 | 0.71 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr6_+_126277842 | 0.71 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr3_+_99536663 | 0.71 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr1_-_159684371 | 0.71 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr1_-_157811588 | 0.70 |
ENST00000368174.4
|
CD5L
|
CD5 molecule-like |
| chr18_+_77160282 | 0.70 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr7_+_113091127 | 0.69 |
ENST00000433518.1
|
TSRM
|
Uncharacterized protein; Zinc finger domain-related protein TSRM |
| chr12_-_772901 | 0.68 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr12_-_109219937 | 0.68 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr15_-_55657428 | 0.68 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr18_+_31185530 | 0.68 |
ENST00000586327.1
|
ASXL3
|
additional sex combs like 3 (Drosophila) |
| chr4_-_80994619 | 0.67 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr8_+_125860939 | 0.66 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr2_+_157292933 | 0.66 |
ENST00000540309.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr5_-_115872142 | 0.66 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 14.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 3.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.6 | 5.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.6 | 2.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.6 | 2.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.5 | 3.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 2.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 4.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 1.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 1.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.4 | 1.6 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.4 | 2.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 4.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 2.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 1.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.4 | 1.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.3 | 3.9 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.3 | 1.9 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 4.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 1.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.3 | 2.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 1.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 5.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.2 | 0.7 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 0.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.7 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 9.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.6 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 2.0 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.2 | 0.8 | GO:0098923 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.2 | 3.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.5 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.2 | 1.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.0 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.2 | 1.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 3.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 3.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.2 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.7 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.6 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.7 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) lung vasculature development(GO:0060426) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 2.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 4.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.5 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 2.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.4 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.6 | GO:0070417 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) cellular response to cold(GO:0070417) |
| 0.1 | 0.7 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 3.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.6 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.5 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.3 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.2 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 2.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 1.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.7 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 10.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 2.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 5.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 2.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.8 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.6 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 3.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.5 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.5 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 8.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 1.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 2.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 4.6 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.3 | 4.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 1.7 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 4.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 3.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 3.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 3.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 8.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 19.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 4.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.1 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 1.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 4.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 3.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 3.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.6 | 2.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 3.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 1.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 1.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 2.9 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 0.9 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.3 | 0.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.3 | 5.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 3.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 9.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.2 | 1.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 4.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 3.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.7 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.7 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 8.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 5.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.8 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 5.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 2.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 2.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 2.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 10.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.1 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 3.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 5.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 5.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 17.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 5.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 1.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.2 | 5.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 2.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 7.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 4.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 3.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 5.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |