Project
Illumina Body Map 2
Navigation
Downloads
Results for GATA5
Z-value: 1.38
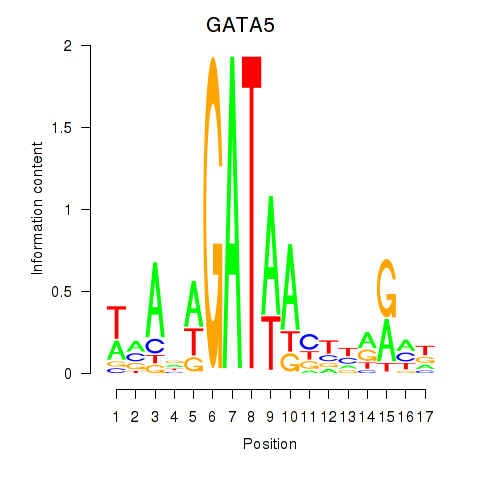
Transcription factors associated with GATA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA5
|
ENSG00000130700.6 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA5 | hg19_v2_chr20_-_61051026_61051057 | -0.28 | 1.2e-01 | Click! |
Activity profile of GATA5 motif
Sorted Z-values of GATA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484155 | 8.73 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484103 | 7.54 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_-_72649763 | 5.77 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr1_+_57320437 | 5.62 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr15_-_54025300 | 4.13 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr21_+_39668478 | 4.01 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_75904918 | 3.91 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_+_75904950 | 3.58 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr2_+_102615416 | 3.08 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr10_+_7745303 | 3.05 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_196857144 | 2.98 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr10_+_7745232 | 2.87 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr9_-_6645628 | 2.74 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr20_-_7238861 | 2.59 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr6_-_135271260 | 2.40 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr6_-_135271219 | 2.39 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chrX_-_55020511 | 2.29 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr8_-_121825088 | 2.21 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr2_-_109605663 | 2.11 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr6_+_78400375 | 2.02 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr11_-_62911693 | 1.90 |
ENST00000417740.1
ENST00000326192.5 |
SLC22A24
|
solute carrier family 22, member 24 |
| chrM_+_3299 | 1.90 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr4_+_110834033 | 1.87 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr19_-_52133588 | 1.84 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr13_-_99910620 | 1.83 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr17_+_6939362 | 1.79 |
ENST00000308027.6
|
SLC16A13
|
solute carrier family 16, member 13 |
| chrM_+_10758 | 1.77 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr8_-_66750978 | 1.77 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr12_+_20968608 | 1.73 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_197300194 | 1.73 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_+_150954493 | 1.66 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr6_-_133055896 | 1.58 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr6_+_53964336 | 1.54 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chr2_+_143635067 | 1.49 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr9_+_6645887 | 1.48 |
ENST00000413145.1
|
RP11-390F4.6
|
RP11-390F4.6 |
| chr14_-_24553834 | 1.47 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr21_-_31864275 | 1.41 |
ENST00000334063.4
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr16_+_82068585 | 1.39 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr6_-_86099898 | 1.37 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr12_-_110883346 | 1.35 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr6_+_140175987 | 1.35 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr13_+_32313658 | 1.33 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr10_-_98031265 | 1.32 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr1_-_159505842 | 1.32 |
ENST00000334857.2
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5 |
| chr14_+_35591020 | 1.28 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr11_+_22694123 | 1.27 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr12_+_10331605 | 1.27 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr1_-_17445930 | 1.25 |
ENST00000375486.4
ENST00000375481.1 ENST00000444885.2 |
PADI2
|
peptidyl arginine deiminase, type II |
| chr6_-_133079022 | 1.24 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr19_-_10450328 | 1.22 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_+_82068873 | 1.21 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr10_+_5090940 | 1.19 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr8_-_6875778 | 1.19 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr5_+_145316120 | 1.15 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr14_+_39735411 | 1.15 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr8_-_6837602 | 1.15 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chrM_+_10464 | 1.10 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_207277590 | 1.10 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr15_-_49913126 | 1.10 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr6_-_4347271 | 1.05 |
ENST00000437430.2
|
RP3-527G5.1
|
RP3-527G5.1 |
| chr17_+_1633755 | 1.05 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr14_+_22928070 | 1.04 |
ENST00000390476.1
|
TRDJ3
|
T cell receptor delta joining 3 |
| chr14_+_21785693 | 1.03 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr12_+_9102632 | 1.02 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr12_-_14967095 | 1.00 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr6_-_52710893 | 0.97 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr15_+_99791716 | 0.96 |
ENST00000558172.1
ENST00000561276.1 ENST00000331450.5 |
LRRC28
|
leucine rich repeat containing 28 |
| chr14_+_22995812 | 0.96 |
ENST00000390520.1
|
TRAJ17
|
T cell receptor alpha joining 17 |
| chr11_+_121461097 | 0.96 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr2_+_102927962 | 0.95 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr7_+_26332645 | 0.95 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr2_+_102928009 | 0.94 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr6_-_24936170 | 0.94 |
ENST00000538035.1
|
FAM65B
|
family with sequence similarity 65, member B |
| chr15_-_49912944 | 0.93 |
ENST00000559905.1
|
FAM227B
|
family with sequence similarity 227, member B |
| chr17_-_41277467 | 0.90 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr3_+_169755919 | 0.90 |
ENST00000492492.1
|
GPR160
|
G protein-coupled receptor 160 |
| chr1_+_174844645 | 0.89 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_10634158 | 0.89 |
ENST00000379591.3
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr2_+_201997676 | 0.86 |
ENST00000462763.1
ENST00000479953.2 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr8_+_20831460 | 0.85 |
ENST00000522604.1
|
RP11-421P23.1
|
RP11-421P23.1 |
| chr3_+_169755715 | 0.85 |
ENST00000355897.5
|
GPR160
|
G protein-coupled receptor 160 |
| chr4_-_83765613 | 0.85 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr6_+_31916733 | 0.84 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_-_157670647 | 0.83 |
ENST00000368184.3
|
FCRL3
|
Fc receptor-like 3 |
| chr19_-_14889349 | 0.82 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr6_+_10633993 | 0.82 |
ENST00000417671.1
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr14_+_22475742 | 0.81 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr4_-_47983519 | 0.80 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr22_+_21213771 | 0.80 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr11_-_61734599 | 0.78 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr2_-_191115229 | 0.78 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_-_142251148 | 0.77 |
ENST00000390360.3
|
TRBV6-4
|
T cell receptor beta variable 6-4 |
| chr13_-_99910673 | 0.76 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr4_-_65275100 | 0.76 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr6_-_55739542 | 0.76 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr7_-_123197733 | 0.76 |
ENST00000470123.1
ENST00000471770.1 |
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr1_+_207277632 | 0.76 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr8_+_93895865 | 0.75 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr1_-_11918988 | 0.75 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr10_-_14596140 | 0.74 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr4_+_108815402 | 0.74 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr3_-_182817297 | 0.74 |
ENST00000539926.1
ENST00000476176.1 |
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr18_+_21032781 | 0.73 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr1_+_62439037 | 0.73 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_+_115572415 | 0.72 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr6_+_106988986 | 0.71 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr9_-_35650900 | 0.70 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr8_+_132952112 | 0.70 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr11_-_72070206 | 0.69 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_+_78245466 | 0.69 |
ENST00000477627.2
|
FAM73A
|
family with sequence similarity 73, member A |
| chr1_+_196743943 | 0.69 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr3_-_51813009 | 0.68 |
ENST00000398780.3
|
IQCF6
|
IQ motif containing F6 |
| chr14_-_22103096 | 0.67 |
ENST00000542433.1
|
OR10G2
|
olfactory receptor, family 10, subfamily G, member 2 |
| chr4_-_65275162 | 0.67 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr14_+_67291158 | 0.65 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr7_+_74379083 | 0.63 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr20_+_32254286 | 0.63 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr9_-_98189055 | 0.62 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr6_-_53409890 | 0.62 |
ENST00000229416.6
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit |
| chr9_-_123239632 | 0.62 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr4_-_48782259 | 0.61 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr8_+_58106143 | 0.61 |
ENST00000521653.1
ENST00000518556.1 |
RP11-513O17.2
|
RP11-513O17.2 |
| chr2_+_182850551 | 0.61 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr8_-_91095099 | 0.60 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr1_+_156123318 | 0.60 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_-_156878482 | 0.60 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr3_+_193311056 | 0.59 |
ENST00000419435.1
|
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chrM_+_10053 | 0.59 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr12_+_40549984 | 0.58 |
ENST00000457989.1
|
AC079630.2
|
AC079630.2 |
| chr7_+_142448053 | 0.57 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr6_-_32908765 | 0.57 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_-_157014865 | 0.57 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr1_-_27226928 | 0.57 |
ENST00000361720.5
|
GPATCH3
|
G patch domain containing 3 |
| chr3_+_193310969 | 0.57 |
ENST00000392437.1
|
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr11_+_128563652 | 0.57 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr14_+_23000026 | 0.55 |
ENST00000390524.1
|
TRAJ13
|
T cell receptor alpha joining 13 |
| chr15_+_99791567 | 0.55 |
ENST00000558879.1
ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28
|
leucine rich repeat containing 28 |
| chr14_+_22392209 | 0.55 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr17_+_36908984 | 0.54 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr22_-_21213676 | 0.53 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr3_+_193311105 | 0.52 |
ENST00000392436.2
|
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr2_+_62132781 | 0.52 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr2_-_47143160 | 0.51 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_-_40170506 | 0.51 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr22_+_17956618 | 0.51 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr10_-_114206649 | 0.50 |
ENST00000369404.3
ENST00000369405.3 |
ZDHHC6
|
zinc finger, DHHC-type containing 6 |
| chr7_-_55583740 | 0.50 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr9_-_70490107 | 0.49 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr12_-_57882577 | 0.49 |
ENST00000393797.2
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr2_-_122247879 | 0.49 |
ENST00000545861.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr5_-_58652788 | 0.48 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_+_133615169 | 0.48 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chrX_+_79675965 | 0.48 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr16_-_28503357 | 0.47 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr15_+_100106244 | 0.47 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr3_+_177545563 | 0.46 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr3_+_46283916 | 0.46 |
ENST00000395940.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr17_-_45908875 | 0.46 |
ENST00000351111.2
ENST00000414011.1 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr1_+_10292308 | 0.45 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr8_+_27629459 | 0.45 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr13_+_108870714 | 0.45 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr3_+_46283944 | 0.45 |
ENST00000452454.1
ENST00000457243.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr2_+_227771404 | 0.45 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr11_+_110225855 | 0.45 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr3_+_46283863 | 0.45 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr3_+_193310918 | 0.45 |
ENST00000361908.3
ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr7_+_16793160 | 0.44 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr2_-_197226875 | 0.44 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_+_147774275 | 0.44 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_+_201997595 | 0.44 |
ENST00000470178.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr9_-_107289490 | 0.43 |
ENST00000277216.3
|
OR13C4
|
olfactory receptor, family 13, subfamily C, member 4 |
| chr4_-_88244010 | 0.43 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_-_160681593 | 0.43 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr17_+_41322483 | 0.42 |
ENST00000341165.6
ENST00000586650.1 ENST00000422280.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr5_-_150537279 | 0.42 |
ENST00000517486.1
ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6
|
annexin A6 |
| chr14_-_23103716 | 0.42 |
ENST00000540461.1
|
OR6J1
|
olfactory receptor, family 6, subfamily J, member 1 (gene/pseudogene) |
| chr12_+_54892550 | 0.41 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr4_-_88244049 | 0.41 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr3_-_108476231 | 0.41 |
ENST00000295755.6
|
RETNLB
|
resistin like beta |
| chr11_-_14379997 | 0.40 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr3_+_108541608 | 0.40 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_-_100575781 | 0.39 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr1_-_100643765 | 0.39 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr11_-_33891362 | 0.38 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_15548591 | 0.38 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr14_+_62164340 | 0.38 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr2_-_128642434 | 0.38 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1-like |
| chr5_+_169758393 | 0.38 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr14_+_61995722 | 0.37 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr8_-_42360015 | 0.37 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr2_+_201997492 | 0.37 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr10_+_15085918 | 0.37 |
ENST00000378225.1
|
OLAH
|
oleoyl-ACP hydrolase |
| chr15_+_45028753 | 0.37 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_-_17380630 | 0.36 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr15_+_100106155 | 0.36 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr6_-_117150198 | 0.36 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr6_-_154677900 | 0.36 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_-_224810070 | 0.36 |
ENST00000429915.1
ENST00000233055.4 |
WDFY1
|
WD repeat and FYVE domain containing 1 |
| chr4_-_175204765 | 0.35 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr15_-_34628951 | 0.35 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 16.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.8 | 3.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.6 | 2.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.5 | 2.7 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 4.8 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 2.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.4 | 1.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 | 1.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 1.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.4 | 1.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 2.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 2.3 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.3 | 1.0 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.3 | 1.3 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.3 | 0.9 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.3 | 1.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.3 | 7.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 1.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.9 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 6.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.8 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 1.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.6 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 4.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.6 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.2 | 1.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.3 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.2 | 5.8 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 2.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 0.6 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 2.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 1.7 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.1 | 0.5 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 1.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 2.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.6 | GO:1990748 | detoxification(GO:0098754) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.4 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 1.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 6.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 5.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.6 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 4.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 1.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:1904430 | cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 1.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.5 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 16.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.9 | 2.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.6 | 5.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 0.9 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.2 | 0.8 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.2 | 2.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 1.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.2 | 0.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.8 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.7 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.5 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 8.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 6.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 12.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.6 | 1.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.6 | 3.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 7.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 2.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.5 | 2.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 1.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 1.2 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.4 | 4.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 2.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 1.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.6 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 4.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 6.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.6 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 2.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.9 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 16.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 6.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.2 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 1.9 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 0.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 2.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 2.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0004320 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 5.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 6.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 2.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 3.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |