Project
Illumina Body Map 2
Navigation
Downloads
Results for GLIS3
Z-value: 0.73
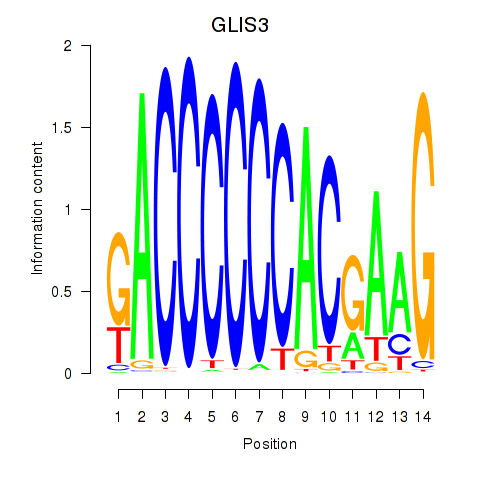
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg19_v2_chr9_-_4299874_4299916 | -0.49 | 4.9e-03 | Click! |
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_57308979 | 2.82 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr2_-_211168332 | 1.95 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr10_+_105314881 | 1.84 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr14_+_105190514 | 1.33 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr19_+_1495362 | 1.16 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr1_+_31886653 | 1.13 |
ENST00000536384.1
|
SERINC2
|
serine incorporator 2 |
| chr21_+_38071430 | 1.08 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr12_+_109577202 | 0.92 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_-_10613862 | 0.87 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr6_+_24495185 | 0.75 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr5_+_17404114 | 0.74 |
ENST00000508677.1
|
RP11-321E2.3
|
RP11-321E2.3 |
| chr6_+_24495067 | 0.71 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr19_-_10613421 | 0.70 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_-_10613361 | 0.66 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr18_+_11751493 | 0.63 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr18_+_11752040 | 0.63 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr15_-_43882140 | 0.62 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr1_-_42801540 | 0.59 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr18_+_11751466 | 0.57 |
ENST00000535121.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr9_-_98279241 | 0.54 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr17_+_80818231 | 0.54 |
ENST00000576996.1
|
TBCD
|
tubulin folding cofactor D |
| chr11_-_118789613 | 0.51 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr11_+_68671310 | 0.50 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr9_-_139658965 | 0.50 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr3_+_127348005 | 0.48 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr12_+_1929421 | 0.48 |
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr12_+_132312931 | 0.47 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr17_+_46018872 | 0.46 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr15_-_43882353 | 0.46 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr1_-_247095236 | 0.45 |
ENST00000478568.1
|
AHCTF1
|
AT hook containing transcription factor 1 |
| chr11_-_299519 | 0.44 |
ENST00000382614.2
|
IFITM5
|
interferon induced transmembrane protein 5 |
| chr5_+_17404147 | 0.44 |
ENST00000507730.1
|
RP11-321E2.3
|
RP11-321E2.3 |
| chr19_-_14224969 | 0.44 |
ENST00000589994.1
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr2_+_219824357 | 0.43 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr20_+_18447771 | 0.43 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr8_-_29120604 | 0.42 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr19_-_56092187 | 0.42 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr2_-_241737128 | 0.41 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr16_+_30006615 | 0.40 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr20_-_44718538 | 0.40 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr3_+_120626919 | 0.39 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr17_-_5015129 | 0.39 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr19_-_48673465 | 0.38 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr9_-_116061476 | 0.38 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr19_+_11909329 | 0.38 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr1_-_42800860 | 0.37 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chr6_+_33172407 | 0.37 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr13_+_25670268 | 0.37 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr20_-_62103862 | 0.36 |
ENST00000344462.4
ENST00000357249.2 ENST00000359125.2 ENST00000360480.3 ENST00000370224.1 ENST00000344425.5 ENST00000354587.3 ENST00000359689.1 |
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2 |
| chr14_+_22931924 | 0.36 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr3_-_160167508 | 0.35 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr3_-_25824925 | 0.33 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr11_+_1463728 | 0.32 |
ENST00000544817.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr17_-_79917645 | 0.32 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr10_+_71389983 | 0.31 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chr11_+_5646213 | 0.31 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr19_-_13213954 | 0.31 |
ENST00000590974.1
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr1_+_59250815 | 0.30 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr3_-_160167301 | 0.29 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr8_+_27183033 | 0.29 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr7_-_50518022 | 0.27 |
ENST00000356889.4
ENST00000420829.1 ENST00000448788.1 ENST00000395556.2 ENST00000422854.1 ENST00000435566.1 ENST00000433017.1 |
FIGNL1
|
fidgetin-like 1 |
| chr8_-_74791051 | 0.27 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr17_+_4675175 | 0.27 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr11_-_8680383 | 0.26 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr3_-_160167540 | 0.26 |
ENST00000496222.1
ENST00000471396.1 ENST00000471155.1 ENST00000309784.4 |
TRIM59
|
tripartite motif containing 59 |
| chr1_+_110527308 | 0.25 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr2_-_241759622 | 0.25 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr8_-_29120580 | 0.25 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr15_+_84116106 | 0.25 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr2_+_220379052 | 0.24 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr1_-_160254913 | 0.24 |
ENST00000440949.3
ENST00000368072.5 ENST00000608310.1 ENST00000556710.1 |
PEX19
DCAF8
DCAF8
|
peroxisomal biogenesis factor 19 DDB1 and CUL4 associated factor 8 DDB1- and CUL4-associated factor 8 |
| chr1_-_53018654 | 0.24 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr8_+_118147498 | 0.23 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr16_-_3285144 | 0.23 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr17_-_3819751 | 0.23 |
ENST00000225538.3
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr8_+_11627205 | 0.22 |
ENST00000455213.2
ENST00000403422.3 ENST00000528323.1 ENST00000284503.6 |
NEIL2
|
nei endonuclease VIII-like 2 (E. coli) |
| chr7_+_23636992 | 0.22 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr19_+_40697514 | 0.22 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr18_+_3451584 | 0.22 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_+_91478493 | 0.21 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr17_+_48912744 | 0.21 |
ENST00000311378.4
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr5_+_172386419 | 0.21 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr1_-_45988542 | 0.20 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr1_-_53019059 | 0.19 |
ENST00000484723.2
ENST00000524582.1 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr17_-_33446820 | 0.19 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr1_+_1950763 | 0.19 |
ENST00000378585.4
|
GABRD
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr19_+_13875316 | 0.19 |
ENST00000319545.8
ENST00000593245.1 ENST00000040663.6 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr22_-_32022280 | 0.18 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr8_+_11627148 | 0.18 |
ENST00000436750.3
|
NEIL2
|
nei endonuclease VIII-like 2 (E. coli) |
| chr8_-_7220490 | 0.18 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr11_+_393428 | 0.17 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr21_-_46330545 | 0.17 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_-_157108130 | 0.17 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr3_-_25824872 | 0.17 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr5_+_172386517 | 0.16 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr19_-_41859814 | 0.12 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr9_-_130742792 | 0.11 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr8_-_12051576 | 0.09 |
ENST00000524571.2
ENST00000533852.2 ENST00000533513.1 ENST00000448228.2 ENST00000534520.1 ENST00000321602.8 |
FAM86B1
|
family with sequence similarity 86, member B1 |
| chr18_+_3451646 | 0.09 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_+_65958693 | 0.09 |
ENST00000445681.1
ENST00000452565.1 |
GS1-124K5.4
|
GS1-124K5.4 |
| chr1_-_157108266 | 0.09 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr22_-_31742218 | 0.09 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr4_-_84255935 | 0.09 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr22_+_18834324 | 0.08 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr1_+_16062820 | 0.08 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr19_-_48673552 | 0.07 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr22_+_50628999 | 0.07 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr11_-_68671264 | 0.07 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr11_+_134855246 | 0.06 |
ENST00000597621.1
|
AP003062.1
|
CDNA FLJ27342 fis, clone TST02993; Uncharacterized protein |
| chr17_-_73178599 | 0.05 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr22_-_50964849 | 0.05 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr5_+_157098534 | 0.05 |
ENST00000409999.3
|
C5orf52
|
chromosome 5 open reading frame 52 |
| chrX_+_118370288 | 0.04 |
ENST00000535419.1
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr11_+_68451943 | 0.02 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chrX_+_118370211 | 0.02 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr12_-_57400227 | 0.02 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr14_+_77228532 | 0.02 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr7_-_127032114 | 0.01 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr5_-_180671172 | 0.01 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr19_+_46850251 | 0.01 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr5_+_41925325 | 0.01 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.5 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 1.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 0.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.5 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.1 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.4 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.5 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 1.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 2.8 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.3 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0075528 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 1.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 2.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.5 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.2 | 0.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 1.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.5 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |