Project
Illumina Body Map 2
Navigation
Downloads
Results for GSC_GSC2
Z-value: 1.03
Transcription factors associated with GSC_GSC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
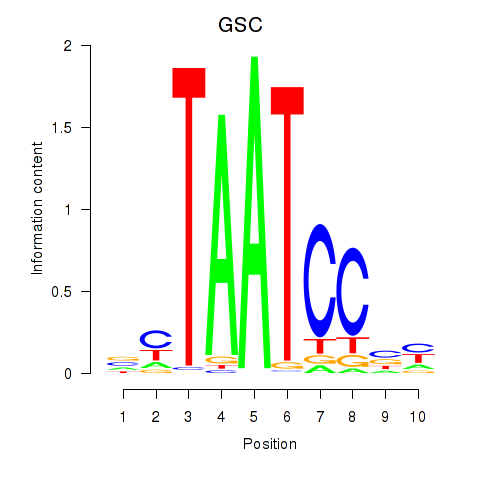
GSC
|
ENSG00000133937.3 | goosecoid homeobox |
|
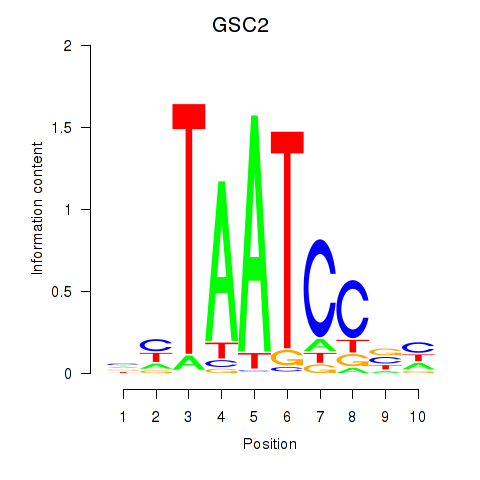
GSC2
|
ENSG00000063515.2 | goosecoid homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GSC | hg19_v2_chr14_-_95236551_95236576 | -0.39 | 2.8e-02 | Click! |
| GSC2 | hg19_v2_chr22_-_19137796_19137796 | 0.38 | 3.0e-02 | Click! |
Activity profile of GSC_GSC2 motif
Sorted Z-values of GSC_GSC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_61522844 | 5.53 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr15_-_27184664 | 4.68 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr17_+_68100989 | 3.98 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_+_169079823 | 3.95 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_+_68101117 | 3.84 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_+_171640291 | 3.36 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr11_+_33563821 | 3.08 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr4_-_16077741 | 2.91 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr11_-_62783303 | 2.49 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr10_-_104178857 | 2.48 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr1_+_151682909 | 2.44 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr1_+_200863949 | 2.44 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr3_-_179691866 | 2.41 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr10_-_49482907 | 2.39 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr13_-_62001982 | 2.37 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chrX_-_6146876 | 2.31 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr11_-_62783276 | 2.30 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr2_+_120770581 | 2.27 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chrX_+_100333709 | 2.23 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr2_+_120770645 | 2.23 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr18_-_35145728 | 2.19 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr9_+_103204553 | 2.17 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_-_47655686 | 2.11 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr18_-_40857493 | 2.05 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr11_-_62752162 | 2.01 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr18_-_64271316 | 1.98 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr6_+_34433844 | 1.98 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chrX_-_54384425 | 1.92 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr18_-_64271363 | 1.90 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr20_+_34556488 | 1.88 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr9_+_12693336 | 1.76 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr3_-_179692042 | 1.75 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr18_-_35145689 | 1.73 |
ENST00000591287.1
ENST00000601019.1 ENST00000601392.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr15_+_54901540 | 1.71 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr16_+_81272287 | 1.66 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr17_-_38859996 | 1.61 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr2_-_175711133 | 1.60 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr2_+_120770686 | 1.55 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_-_33984741 | 1.55 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chrX_-_138724677 | 1.54 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr3_-_10547192 | 1.54 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr12_+_78224667 | 1.53 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr11_+_33563618 | 1.50 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr2_+_173792893 | 1.46 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr3_-_19975665 | 1.44 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr12_+_15125954 | 1.41 |
ENST00000266395.2
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr14_+_96722152 | 1.38 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr18_-_35145593 | 1.31 |
ENST00000334919.5
ENST00000591282.1 ENST00000588597.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr18_-_24443151 | 1.28 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chrX_-_24665353 | 1.28 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr5_+_161274685 | 1.27 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr14_-_101034811 | 1.26 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr3_-_65583561 | 1.24 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_+_161275320 | 1.24 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr18_-_40857447 | 1.24 |
ENST00000590752.1
ENST00000596867.1 |
SYT4
|
synaptotagmin IV |
| chr5_+_161274940 | 1.24 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_170626418 | 1.23 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr16_-_68269971 | 1.20 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr14_-_70040339 | 1.19 |
ENST00000599174.1
|
CCDC177
|
Homo sapiens coiled-coil domain containing 177 (CCDC177), mRNA. |
| chr11_-_128712362 | 1.18 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr10_-_86001210 | 1.16 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr12_-_113574028 | 1.16 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr20_-_39995467 | 1.14 |
ENST00000332312.3
|
EMILIN3
|
elastin microfibril interfacer 3 |
| chr1_+_20396649 | 1.14 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chrX_-_24665208 | 1.13 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_-_40555176 | 1.11 |
ENST00000338305.6
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr11_-_62752455 | 1.07 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_62752429 | 1.05 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr15_+_48498480 | 1.02 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chrX_-_138724994 | 1.00 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr17_-_19651668 | 0.98 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_-_77272765 | 0.97 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr11_-_35440579 | 0.96 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_35440796 | 0.95 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr17_+_48911942 | 0.94 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr16_+_87636474 | 0.93 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr10_-_74714533 | 0.92 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chr1_+_197237352 | 0.92 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr8_+_65285851 | 0.90 |
ENST00000520799.1
ENST00000521441.1 |
LINC00966
|
long intergenic non-protein coding RNA 966 |
| chr16_+_4421841 | 0.87 |
ENST00000304735.3
|
VASN
|
vasorin |
| chr5_+_5140436 | 0.87 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr17_+_4643337 | 0.87 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr3_-_98241358 | 0.85 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr13_+_114321463 | 0.84 |
ENST00000335678.6
|
GRK1
|
G protein-coupled receptor kinase 1 |
| chr18_-_40857329 | 0.82 |
ENST00000593720.1
|
SYT4
|
synaptotagmin IV |
| chr10_+_18240834 | 0.82 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr7_-_138482849 | 0.80 |
ENST00000353492.4
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr3_+_49058444 | 0.78 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr10_+_102222798 | 0.77 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr17_+_44668387 | 0.76 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chrX_-_10588595 | 0.75 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr9_-_13279563 | 0.74 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr7_-_138482933 | 0.74 |
ENST00000310018.2
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4 |
| chr10_+_18240753 | 0.74 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr17_+_4643300 | 0.73 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr10_+_18240814 | 0.73 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr6_-_41130914 | 0.73 |
ENST00000373113.3
ENST00000338469.3 |
TREM2
|
triggering receptor expressed on myeloid cells 2 |
| chr2_+_189156586 | 0.72 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr12_-_49523896 | 0.72 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr3_-_170626376 | 0.71 |
ENST00000487522.1
ENST00000474417.1 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr2_-_27486951 | 0.68 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr2_+_189156721 | 0.68 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr12_+_119772734 | 0.68 |
ENST00000539847.1
|
CCDC60
|
coiled-coil domain containing 60 |
| chr14_-_101034407 | 0.67 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr3_-_193272874 | 0.67 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr12_-_52604607 | 0.67 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr9_-_13279589 | 0.67 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr15_-_52483566 | 0.66 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr8_+_55528627 | 0.66 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr22_-_19137796 | 0.64 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr8_-_114449112 | 0.62 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr2_+_189156389 | 0.61 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_+_63050785 | 0.61 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr17_-_40264692 | 0.60 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr22_-_25342662 | 0.60 |
ENST00000407886.1
|
TMEM211
|
transmembrane protein 211 |
| chr5_-_16738451 | 0.60 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr6_-_41130841 | 0.60 |
ENST00000373122.4
|
TREM2
|
triggering receptor expressed on myeloid cells 2 |
| chr5_+_140868717 | 0.59 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr3_-_193272741 | 0.58 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr2_-_30144432 | 0.58 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr7_+_8474817 | 0.57 |
ENST00000429542.1
|
NXPH1
|
neurexophilin 1 |
| chr1_+_171939240 | 0.57 |
ENST00000523513.1
|
DNM3
|
dynamin 3 |
| chr17_-_19651654 | 0.57 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_193272588 | 0.56 |
ENST00000295548.3
|
ATP13A4
|
ATPase type 13A4 |
| chr18_+_32290218 | 0.56 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr3_+_111393659 | 0.56 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_111794446 | 0.55 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr17_+_44668035 | 0.55 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_+_6949964 | 0.54 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr11_-_76155700 | 0.53 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr7_-_78400364 | 0.51 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_-_146461027 | 0.51 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_201140673 | 0.51 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr5_-_33984786 | 0.50 |
ENST00000296589.4
|
SLC45A2
|
solute carrier family 45, member 2 |
| chr12_-_13248562 | 0.49 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr7_+_8474693 | 0.49 |
ENST00000438764.1
|
NXPH1
|
neurexophilin 1 |
| chr6_+_121756809 | 0.49 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chrX_+_65382433 | 0.49 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr16_+_58010339 | 0.49 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr7_-_96132835 | 0.49 |
ENST00000356686.1
|
C7orf76
|
chromosome 7 open reading frame 76 |
| chr8_+_70378852 | 0.48 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr2_-_29297127 | 0.48 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr8_-_37457350 | 0.47 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr1_+_114522049 | 0.46 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr11_-_40315640 | 0.46 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr5_+_173930710 | 0.45 |
ENST00000511707.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr22_-_38480100 | 0.44 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr6_+_138266498 | 0.43 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr9_+_17134980 | 0.43 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr13_+_36050881 | 0.43 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr11_-_76155618 | 0.42 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_48207115 | 0.42 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr18_-_31628558 | 0.42 |
ENST00000535384.1
|
NOL4
|
nucleolar protein 4 |
| chr6_-_154568551 | 0.41 |
ENST00000519190.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr12_-_13248732 | 0.39 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chrX_+_139791917 | 0.39 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr2_+_128175997 | 0.38 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr17_-_4643161 | 0.38 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr7_-_84816122 | 0.38 |
ENST00000444867.1
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr12_-_13248705 | 0.37 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chrX_+_65382381 | 0.37 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chrX_+_41306575 | 0.36 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr11_-_111783595 | 0.35 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr13_-_95131923 | 0.33 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr12_-_13248598 | 0.33 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr11_+_20044096 | 0.33 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr7_-_128415844 | 0.33 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr2_-_65659762 | 0.33 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr6_+_31588478 | 0.30 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr17_-_40021656 | 0.30 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr19_+_2476116 | 0.30 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr20_+_33292507 | 0.30 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_26633480 | 0.30 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr8_-_87755878 | 0.29 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr7_+_138482695 | 0.29 |
ENST00000422794.2
ENST00000397602.3 ENST00000442682.2 ENST00000458494.1 ENST00000413208.1 |
TMEM213
|
transmembrane protein 213 |
| chr7_-_155326532 | 0.28 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
| chr4_+_159727222 | 0.28 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr16_-_54304608 | 0.28 |
ENST00000561336.1
|
RP11-324D17.1
|
HCG2045435; Uncharacterized protein |
| chr15_-_31453157 | 0.28 |
ENST00000559177.1
ENST00000558445.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr17_-_26903900 | 0.27 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr7_-_38969150 | 0.26 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr15_+_44092784 | 0.25 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chrX_-_124097620 | 0.25 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr7_+_129847688 | 0.25 |
ENST00000297819.3
|
SSMEM1
|
serine-rich single-pass membrane protein 1 |
| chr15_+_33603147 | 0.25 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr12_+_7053172 | 0.24 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr7_+_93535866 | 0.24 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chrX_+_69509927 | 0.23 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chrX_-_10588459 | 0.23 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_+_192141611 | 0.23 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr1_-_43638168 | 0.23 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr6_-_154567984 | 0.23 |
ENST00000517438.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_111393501 | 0.22 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr6_-_154568815 | 0.21 |
ENST00000519344.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr20_-_18774614 | 0.21 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr9_+_17135016 | 0.21 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr19_-_46142362 | 0.21 |
ENST00000586770.1
ENST00000591721.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_57967854 | 0.21 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr11_-_119217365 | 0.20 |
ENST00000360167.4
ENST00000555262.1 ENST00000449574.2 ENST00000445041.2 |
MFRP
C1QTNF5
|
membrane frizzled-related protein C1q and tumor necrosis factor related protein 5 |
| chr12_+_7053228 | 0.20 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GSC_GSC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 1.4 | 4.1 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 1.2 | 6.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.6 | 1.8 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.6 | 2.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.6 | 4.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.5 | 2.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 | 1.9 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.5 | 7.5 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.5 | 5.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 1.3 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.4 | 4.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 8.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 1.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 2.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 1.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 2.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 1.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 9.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 1.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 2.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 4.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 2.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.7 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 1.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 6.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.4 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 2.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.9 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.9 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.3 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 1.7 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 8.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 4.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 4.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 2.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 9.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.0 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 8.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 4.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.5 | 2.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 8.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 2.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 1.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 1.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.4 | 4.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 4.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 7.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 0.8 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.2 | 6.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 0.7 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.2 | 1.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 1.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 4.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 4.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 2.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 7.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.8 | GO:0004871 | signal transducer activity(GO:0004871) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 4.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 8.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 8.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 5.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 2.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |