Project
Illumina Body Map 2
Navigation
Downloads
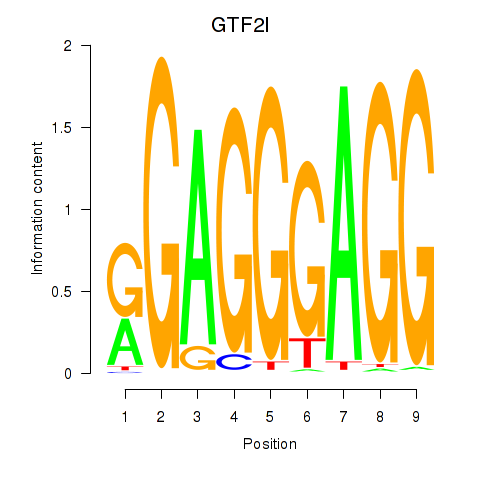
Results for GTF2I
Z-value: 1.95
Transcription factors associated with GTF2I
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GTF2I
|
ENSG00000077809.8 | general transcription factor IIi |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GTF2I | hg19_v2_chr7_+_74072288_74072357 | 0.25 | 1.7e-01 | Click! |
Activity profile of GTF2I motif
Sorted Z-values of GTF2I motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_203444887 | 6.73 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chr14_+_24837226 | 6.17 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr12_-_16759440 | 4.34 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_13044787 | 4.16 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr9_-_34589734 | 3.93 |
ENST00000378980.3
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr12_-_16759711 | 3.74 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr11_+_125034586 | 3.74 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr9_-_34589700 | 3.60 |
ENST00000351266.4
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr14_-_61191049 | 3.35 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr11_+_125034640 | 3.29 |
ENST00000542175.1
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr12_-_115121962 | 3.24 |
ENST00000349155.2
|
TBX3
|
T-box 3 |
| chr4_-_16900242 | 3.12 |
ENST00000502640.1
ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr15_-_37392703 | 3.09 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr4_-_16900410 | 3.08 |
ENST00000304523.5
|
LDB2
|
LIM domain binding 2 |
| chr17_-_74533734 | 3.05 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr11_+_394196 | 2.95 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr16_-_65155979 | 2.94 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr5_-_121413974 | 2.92 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr7_+_73442487 | 2.92 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr3_-_168864427 | 2.87 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr15_-_37392724 | 2.83 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr9_+_109625378 | 2.79 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr14_-_27066636 | 2.77 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr7_+_73442422 | 2.77 |
ENST00000358929.4
ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN
|
elastin |
| chr17_+_60704762 | 2.69 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr16_-_65155833 | 2.64 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chrX_-_119445306 | 2.63 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr4_-_16900217 | 2.60 |
ENST00000441778.2
|
LDB2
|
LIM domain binding 2 |
| chr7_+_30951461 | 2.57 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr7_+_73442457 | 2.55 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr12_+_58005204 | 2.54 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chrX_-_133119476 | 2.53 |
ENST00000543339.1
|
GPC3
|
glypican 3 |
| chr17_-_74533963 | 2.51 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr2_-_19558373 | 2.49 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr1_+_153651078 | 2.43 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr2_-_218867711 | 2.43 |
ENST00000446903.1
|
TNS1
|
tensin 1 |
| chrX_-_133119670 | 2.40 |
ENST00000394299.2
|
GPC3
|
glypican 3 |
| chr17_+_37783170 | 2.35 |
ENST00000254079.4
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr17_-_15165854 | 2.34 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr17_+_37783197 | 2.33 |
ENST00000582680.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_+_86046433 | 2.32 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr17_+_37783453 | 2.31 |
ENST00000579000.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chrX_-_133119895 | 2.27 |
ENST00000370818.3
|
GPC3
|
glypican 3 |
| chr4_-_16900184 | 2.26 |
ENST00000515064.1
|
LDB2
|
LIM domain binding 2 |
| chr14_-_54423529 | 2.26 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr19_-_49243845 | 2.25 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr12_+_175930 | 2.25 |
ENST00000538872.1
ENST00000326261.4 |
IQSEC3
|
IQ motif and Sec7 domain 3 |
| chr2_+_189156586 | 2.25 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_189156721 | 2.24 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr16_-_65156235 | 2.23 |
ENST00000564317.1
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr11_-_72385437 | 2.22 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chrX_-_119445263 | 2.21 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr1_-_119530428 | 2.21 |
ENST00000369429.3
|
TBX15
|
T-box 15 |
| chr4_+_106816644 | 2.20 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chr22_-_38380543 | 2.20 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr7_-_27170352 | 2.18 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr6_+_19837592 | 2.18 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr17_+_37782955 | 2.17 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr3_-_64673668 | 2.15 |
ENST00000498707.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr3_-_64673289 | 2.14 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr1_+_157963391 | 2.13 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr17_-_15165825 | 2.12 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr2_-_56150910 | 2.12 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr22_+_45898712 | 2.11 |
ENST00000455233.1
ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1
|
fibulin 1 |
| chr3_-_64673656 | 2.11 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr9_+_102584128 | 2.10 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr12_-_8088773 | 2.09 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr9_-_34590121 | 2.07 |
ENST00000417345.1
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr5_-_44388899 | 2.05 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chrX_+_144899314 | 2.02 |
ENST00000335565.4
|
SLITRK2
|
SLIT and NTRK-like family, member 2 |
| chr2_+_176995011 | 2.02 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr7_+_31092109 | 2.02 |
ENST00000409363.1
|
ADCYAP1R1
|
adenylate cyclase activating polypeptide 1 (pituitary) receptor type I |
| chr6_+_43739697 | 2.01 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr19_+_35629702 | 2.00 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr13_-_36705425 | 1.99 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr14_-_21562648 | 1.95 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr1_+_157963063 | 1.95 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr11_+_7506713 | 1.95 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr18_-_45935663 | 1.91 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr1_-_147245445 | 1.91 |
ENST00000430508.1
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr20_+_43374421 | 1.91 |
ENST00000372861.3
|
KCNK15
|
potassium channel, subfamily K, member 15 |
| chr4_-_177713788 | 1.90 |
ENST00000280193.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr2_+_189156389 | 1.89 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr1_-_147245484 | 1.87 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr14_+_37131058 | 1.86 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chrX_+_144899416 | 1.86 |
ENST00000447897.2
|
SLITRK2
|
SLIT and NTRK-like family, member 2 |
| chr19_-_46272462 | 1.84 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr19_+_55999916 | 1.83 |
ENST00000587166.1
ENST00000389623.6 |
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr3_-_64431058 | 1.83 |
ENST00000564377.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr5_-_146833222 | 1.83 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr8_-_25902876 | 1.83 |
ENST00000520164.1
|
EBF2
|
early B-cell factor 2 |
| chr19_+_35630022 | 1.82 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_-_24715478 | 1.82 |
ENST00000456299.2
ENST00000540811.1 |
RP11-444D3.1
|
RP11-444D3.1 |
| chr14_-_61190754 | 1.82 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr2_+_42275153 | 1.82 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr11_+_7506837 | 1.81 |
ENST00000528758.1
|
OLFML1
|
olfactomedin-like 1 |
| chr22_-_30642782 | 1.81 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr2_+_85981008 | 1.80 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr3_-_73673991 | 1.78 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr7_+_31092076 | 1.78 |
ENST00000304166.4
|
ADCYAP1R1
|
adenylate cyclase activating polypeptide 1 (pituitary) receptor type I |
| chr18_+_46065570 | 1.78 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr1_-_68299130 | 1.77 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr14_-_21562671 | 1.76 |
ENST00000554923.1
|
ZNF219
|
zinc finger protein 219 |
| chr17_-_40828969 | 1.76 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr5_+_149980622 | 1.75 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr12_-_16760021 | 1.74 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr7_+_73442102 | 1.73 |
ENST00000445912.1
ENST00000252034.7 |
ELN
|
elastin |
| chr19_+_18496957 | 1.73 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr6_-_6007200 | 1.73 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr6_-_75915757 | 1.71 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr11_-_123065989 | 1.69 |
ENST00000448775.2
|
CLMP
|
CXADR-like membrane protein |
| chr9_-_129885010 | 1.68 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr1_-_72748140 | 1.68 |
ENST00000434200.1
|
NEGR1
|
neuronal growth regulator 1 |
| chr1_-_214724566 | 1.68 |
ENST00000366956.5
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr5_-_146833485 | 1.65 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_133748913 | 1.65 |
ENST00000310926.4
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr2_-_227664474 | 1.64 |
ENST00000305123.5
|
IRS1
|
insulin receptor substrate 1 |
| chr9_-_129884902 | 1.64 |
ENST00000373417.1
|
ANGPTL2
|
angiopoietin-like 2 |
| chr7_+_65338312 | 1.63 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr8_-_93107827 | 1.63 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_36904437 | 1.63 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr17_+_1959369 | 1.61 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr7_+_65338230 | 1.61 |
ENST00000360768.3
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr17_+_7308172 | 1.60 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr7_-_19157248 | 1.60 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr8_-_38325219 | 1.60 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr10_-_88729200 | 1.60 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr6_-_29600559 | 1.60 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_-_132262060 | 1.59 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr3_+_139654018 | 1.59 |
ENST00000458420.3
|
CLSTN2
|
calsyntenin 2 |
| chr12_-_16758873 | 1.58 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr7_-_32111009 | 1.58 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr8_+_19796381 | 1.58 |
ENST00000524029.1
ENST00000522701.1 ENST00000311322.8 |
LPL
|
lipoprotein lipase |
| chr9_+_87284622 | 1.57 |
ENST00000395882.1
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chrX_-_110039038 | 1.57 |
ENST00000372042.1
ENST00000482160.1 ENST00000444321.2 ENST00000218054.4 |
CHRDL1
|
chordin-like 1 |
| chr19_-_15311713 | 1.57 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr12_-_16760195 | 1.56 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_-_168864315 | 1.56 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_+_114522049 | 1.56 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr12_+_49209348 | 1.55 |
ENST00000536187.2
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr8_-_22549856 | 1.55 |
ENST00000522910.1
|
EGR3
|
early growth response 3 |
| chr8_-_22550815 | 1.54 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr12_-_16758835 | 1.54 |
ENST00000541295.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_+_20255123 | 1.53 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr1_+_13910757 | 1.52 |
ENST00000376061.4
ENST00000513143.1 |
PDPN
|
podoplanin |
| chr2_+_27301435 | 1.52 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr17_-_40829026 | 1.51 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr12_-_6484376 | 1.51 |
ENST00000360168.3
ENST00000358945.3 |
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr1_+_164528866 | 1.49 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr19_-_54984354 | 1.48 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr12_-_6484715 | 1.48 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chrX_-_110038990 | 1.46 |
ENST00000372045.1
ENST00000394797.4 |
CHRDL1
|
chordin-like 1 |
| chr10_-_88729069 | 1.46 |
ENST00000609457.1
|
MMRN2
|
multimerin 2 |
| chr18_+_46065483 | 1.46 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr6_-_112575687 | 1.46 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr5_+_92919043 | 1.45 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr7_+_73245193 | 1.44 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr4_+_41540160 | 1.43 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_53228079 | 1.43 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr12_+_54519842 | 1.43 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr19_+_55587266 | 1.41 |
ENST00000201647.6
ENST00000540810.1 |
EPS8L1
|
EPS8-like 1 |
| chr6_+_1389989 | 1.41 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr22_+_38609538 | 1.40 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr9_+_133971909 | 1.40 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr16_-_77468945 | 1.39 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr11_+_12399071 | 1.39 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr11_-_62689046 | 1.39 |
ENST00000306960.3
ENST00000543973.1 |
CHRM1
|
cholinergic receptor, muscarinic 1 |
| chr6_-_112575758 | 1.37 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr5_+_140800638 | 1.37 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr19_-_42573650 | 1.36 |
ENST00000593562.1
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5 |
| chr10_+_88718314 | 1.36 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr1_+_201617264 | 1.36 |
ENST00000367296.4
|
NAV1
|
neuron navigator 1 |
| chr17_+_37784749 | 1.35 |
ENST00000394265.1
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr9_+_87284675 | 1.34 |
ENST00000376208.1
ENST00000304053.6 ENST00000277120.3 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr10_+_120967072 | 1.32 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr1_-_208084729 | 1.32 |
ENST00000310833.7
ENST00000356522.4 |
CD34
|
CD34 molecule |
| chr2_+_189156638 | 1.31 |
ENST00000410051.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr6_-_29600832 | 1.31 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr13_+_93879085 | 1.31 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chrX_-_110039286 | 1.31 |
ENST00000434224.1
|
CHRDL1
|
chordin-like 1 |
| chr22_-_36236265 | 1.31 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_+_48133459 | 1.31 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr8_+_15397732 | 1.30 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr3_-_133748758 | 1.30 |
ENST00000493729.1
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr1_+_13910479 | 1.30 |
ENST00000509009.1
|
PDPN
|
podoplanin |
| chr12_-_16761007 | 1.29 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_+_30241995 | 1.29 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr1_+_201617450 | 1.28 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr19_+_45973120 | 1.27 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr8_-_22550691 | 1.27 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr6_-_112575912 | 1.27 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr10_-_14372870 | 1.26 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr10_+_88718397 | 1.26 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr19_-_51071302 | 1.26 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr2_+_239756671 | 1.25 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr2_-_50574856 | 1.25 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr7_+_100770328 | 1.25 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr19_-_46272106 | 1.24 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr19_-_291365 | 1.24 |
ENST00000591572.1
ENST00000269812.3 ENST00000434325.2 |
PPAP2C
|
phosphatidic acid phosphatase type 2C |
| chr11_-_106889250 | 1.24 |
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GTF2I
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.5 | 3.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 1.4 | 9.7 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 1.3 | 5.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 1.3 | 3.8 | GO:0086044 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 1.2 | 3.7 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.2 | 16.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.1 | 3.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 1.1 | 3.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 1.0 | 1.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.0 | 10.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.9 | 2.8 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.9 | 0.9 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.9 | 4.3 | GO:0048749 | compound eye development(GO:0048749) |
| 0.9 | 2.6 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.8 | 2.5 | GO:0072011 | mesangial cell-matrix adhesion(GO:0035759) glomerular endothelium development(GO:0072011) |
| 0.8 | 9.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.8 | 5.6 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.7 | 2.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.7 | 2.1 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.7 | 2.1 | GO:0071336 | bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) secretion by lung epithelial cell involved in lung growth(GO:0061033) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.7 | 1.3 | GO:0048371 | lateral mesodermal cell differentiation(GO:0048371) |
| 0.7 | 2.0 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.6 | 6.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.6 | 5.0 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.6 | 3.6 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.6 | 2.9 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.6 | 2.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.6 | 1.1 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.6 | 1.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.6 | 1.7 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.6 | 1.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.6 | 2.2 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.6 | 2.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.5 | 1.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.5 | 1.6 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.5 | 2.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.5 | 3.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.5 | 0.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.5 | 1.5 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.5 | 1.9 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.5 | 1.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 4.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 2.3 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.5 | 2.3 | GO:0061227 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.4 | 2.7 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.4 | 2.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 | 4.4 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.4 | 0.4 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.4 | 3.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 2.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 1.2 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.4 | 2.4 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.4 | 0.8 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.4 | 0.8 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.4 | 1.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 2.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 2.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.4 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 1.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.4 | 1.1 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.4 | 1.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 1.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.3 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 1.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 3.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 1.3 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) allantois development(GO:1905069) |
| 0.3 | 1.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.3 | 1.6 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 4.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 5.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.9 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.3 | 0.9 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.3 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 6.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 1.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 2.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.3 | 11.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.3 | 0.8 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.3 | 1.6 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 3.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 1.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.3 | 0.3 | GO:0008050 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.3 | 6.0 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.5 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.3 | 1.8 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 0.8 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.3 | 1.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 1.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 2.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.7 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.2 | 2.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.7 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 0.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 2.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.3 | GO:0015853 | adenine transport(GO:0015853) |
| 0.2 | 2.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.7 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.2 | 1.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 0.7 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.2 | 1.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.2 | 0.6 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.2 | 0.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.6 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.2 | 1.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 2.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 1.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 6.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 0.6 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 1.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.2 | 1.0 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 1.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 | 0.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 0.2 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.2 | 4.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.4 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 1.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 | 1.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.5 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.2 | 5.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 4.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 1.1 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 | 2.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 1.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 1.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 1.0 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.2 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 1.0 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 0.8 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.2 | 1.8 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 2.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 1.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.2 | 1.7 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.3 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.6 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.6 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.6 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 1.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.3 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.4 | GO:1990575 | mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.6 | GO:0048807 | ectoderm and mesoderm interaction(GO:0007499) female genitalia morphogenesis(GO:0048807) |
| 0.1 | 1.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.8 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.7 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.7 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.7 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 1.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 4.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.9 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 1.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 1.2 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.1 | 4.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 1.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.0 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 2.5 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.1 | 1.8 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 1.0 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.6 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.9 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 3.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.5 | GO:0060460 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 8.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 2.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.1 | GO:0071505 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.8 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.5 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 2.4 | GO:0097205 | renal filtration(GO:0097205) |
| 0.1 | 1.7 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.9 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 2.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 1.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 7.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 0.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.2 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) GDP metabolic process(GO:0046710) |
| 0.1 | 1.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.7 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.1 | 0.8 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:1990927 | vesicle-mediated cholesterol transport(GO:0090119) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 1.4 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 3.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.4 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 2.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 2.3 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.1 | 0.3 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 1.5 | GO:1901739 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.6 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.1 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 8.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.3 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.9 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 2.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 3.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.3 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.2 | GO:2000611 | thyroid-stimulating hormone secretion(GO:0070460) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.7 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.3 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 2.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 6.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 1.2 | GO:1904261 | positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 1.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 4.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 1.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.5 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.7 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.1 | 0.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.0 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0032352 | positive regulation of hormone metabolic process(GO:0032352) |
| 0.1 | 0.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.5 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.8 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 4.3 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 8.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 1.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 2.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.9 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.5 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.9 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 1.9 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.7 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.6 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 1.4 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0061013 | regulation of mRNA catabolic process(GO:0061013) |
| 0.0 | 0.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 3.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.9 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 1.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 2.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 3.3 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.9 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.4 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 1.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 2.0 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 1.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 1.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.9 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 1.0 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 2.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 2.9 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 1.8 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.5 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 1.1 | 12.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.9 | 2.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.8 | 0.8 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.8 | 3.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 2.2 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.6 | 1.8 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.6 | 1.8 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.6 | 1.7 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.5 | 2.3 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 1.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.4 | 1.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 1.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.4 | 2.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 2.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.3 | 4.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 4.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.7 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 7.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 6.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 5.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 9.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 1.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 11.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 4.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 4.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 5.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 3.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 3.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 47.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.9 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 1.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 12.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 3.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 6.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 1.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 3.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 8.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 2.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 2.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 1.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 16.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 9.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 4.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.5 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.8 | 7.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 1.4 | 5.6 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 1.3 | 3.8 | GO:0086076 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 1.1 | 3.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.9 | 9.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.9 | 2.6 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.8 | 10.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.8 | 3.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.7 | 2.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.7 | 5.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.7 | 3.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 3.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.6 | 3.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.6 | 3.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 2.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.5 | 1.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 1.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.4 | 1.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.4 | 2.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 2.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.4 | 1.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 4.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 1.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 4.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 3.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 2.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 6.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 2.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 1.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 1.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.3 | 2.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 2.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 1.0 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.3 | 1.9 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.3 | 5.4 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.3 | 4.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 1.9 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 3.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.3 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 0.9 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.3 | 1.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 2.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 0.8 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.3 | 0.8 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 1.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.3 | 2.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.7 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.2 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 3.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.9 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 0.6 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.2 | 29.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 0.6 | GO:0030395 | lactose binding(GO:0030395) |
| 0.2 | 0.6 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.2 | 1.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 3.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 4.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.5 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.2 | 2.5 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 1.2 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 1.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 6.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 3.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 0.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 1.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 2.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 1.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 2.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 2.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 2.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.4 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 3.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 3.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 6.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 4.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 1.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.5 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.3 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 5.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 2.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 13.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.5 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 1.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 7.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 6.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 4.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 3.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 11.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 3.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 4.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 9.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.3 | GO:0003823 | antigen binding(GO:0003823) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 12.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 7.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 6.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 6.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 29.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 6.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 6.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 4.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 15.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 10.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 6.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 10.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 6.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 7.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 11.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 11.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 5.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 1.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 9.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 5.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 3.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 3.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.1 | REACTOME S PHASE | Genes involved in S Phase |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME HIV INFECTION | Genes involved in HIV Infection |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |